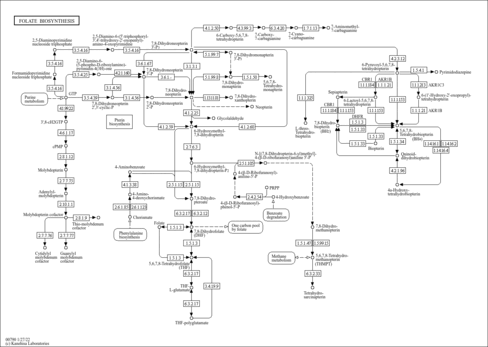
| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Sepiapterin,1TMS,isomer #1 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N)=NC2=O | 2418.8 | Semi standard non polar | 33892256 |
| Sepiapterin,1TMS,isomer #2 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N)=NC2=O | 2469.8 | Semi standard non polar | 33892256 |
| Sepiapterin,1TMS,isomer #3 | C[C@H](O)C(=O)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C)=NC2=O | 2439.3 | Semi standard non polar | 33892256 |
| Sepiapterin,1TMS,isomer #4 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1 | 2287.7 | Semi standard non polar | 33892256 |
| Sepiapterin,1TMS,isomer #5 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O | 2423.3 | Semi standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N)=NC2=O | 2471.5 | Semi standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N)=NC2=O | 2457.3 | Standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N)=NC2=O | 4264.8 | Standard polar | 33892256 |
| Sepiapterin,2TMS,isomer #10 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2300.1 | Semi standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #10 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2569.5 | Standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #10 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 4516.6 | Standard polar | 33892256 |
| Sepiapterin,2TMS,isomer #11 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O | 2298.2 | Semi standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #11 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O | 2443.9 | Standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #11 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O | 4109.8 | Standard polar | 33892256 |
| Sepiapterin,2TMS,isomer #2 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C)=NC2=O | 2395.6 | Semi standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #2 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C)=NC2=O | 2575.5 | Standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #2 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C)=NC2=O | 4774.6 | Standard polar | 33892256 |
| Sepiapterin,2TMS,isomer #3 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O | 2367.2 | Semi standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #3 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O | 2440.9 | Standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #3 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O | 4311.7 | Standard polar | 33892256 |
| Sepiapterin,2TMS,isomer #4 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1 | 2282.5 | Semi standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #4 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1 | 2480.6 | Standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #4 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1 | 4079.9 | Standard polar | 33892256 |
| Sepiapterin,2TMS,isomer #5 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C)=NC2=O | 2468.7 | Semi standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #5 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C)=NC2=O | 2536.1 | Standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #5 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C)=NC2=O | 4808.0 | Standard polar | 33892256 |
| Sepiapterin,2TMS,isomer #6 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O | 2408.2 | Semi standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #6 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O | 2435.6 | Standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #6 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O | 4330.1 | Standard polar | 33892256 |
| Sepiapterin,2TMS,isomer #7 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1 | 2330.3 | Semi standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #7 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1 | 2482.0 | Standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #7 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1 | 4090.0 | Standard polar | 33892256 |
| Sepiapterin,2TMS,isomer #8 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2377.9 | Semi standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #8 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2538.8 | Standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #8 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 4704.5 | Standard polar | 33892256 |
| Sepiapterin,2TMS,isomer #9 | C[C@H](O)C(=O)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2388.1 | Semi standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #9 | C[C@H](O)C(=O)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2548.0 | Standard non polar | 33892256 |
| Sepiapterin,2TMS,isomer #9 | C[C@H](O)C(=O)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4745.2 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C)=NC2=O | 2461.3 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C)=NC2=O | 2561.4 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C)=NC2=O | 4542.0 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #10 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2410.7 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #10 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2600.6 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #10 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 4281.3 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #11 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O | 2358.0 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #11 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O | 2527.1 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #11 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O | 3895.5 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #12 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2400.8 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #12 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2611.7 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #12 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4325.4 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #13 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2329.5 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #13 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2633.9 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #13 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 4120.0 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #14 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2312.4 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #14 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2610.8 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #14 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 4176.7 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #2 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O | 2417.8 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #2 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O | 2493.9 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #2 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O | 4080.0 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #3 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1 | 2345.8 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #3 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1 | 2537.8 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #3 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1 | 3870.1 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #4 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2363.3 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #4 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2605.7 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #4 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 4396.2 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #5 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2405.9 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #5 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2627.1 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #5 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4419.8 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #6 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C([NH]C(N[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2343.7 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #6 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C([NH]C(N[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2613.2 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #6 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C([NH]C(N[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 4259.0 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #7 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O | 2293.3 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #7 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O | 2513.4 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #7 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O | 3894.5 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #8 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2433.5 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #8 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2584.9 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #8 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 4402.8 | Standard polar | 33892256 |
| Sepiapterin,3TMS,isomer #9 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2443.8 | Semi standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #9 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2619.7 | Standard non polar | 33892256 |
| Sepiapterin,3TMS,isomer #9 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4428.9 | Standard polar | 33892256 |
| Sepiapterin,4TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2456.6 | Semi standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2595.5 | Standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 4169.4 | Standard polar | 33892256 |
| Sepiapterin,4TMS,isomer #10 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2424.5 | Semi standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #10 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2672.8 | Standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #10 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 3935.7 | Standard polar | 33892256 |
| Sepiapterin,4TMS,isomer #11 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2389.2 | Semi standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #11 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2675.2 | Standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #11 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 3747.9 | Standard polar | 33892256 |
| Sepiapterin,4TMS,isomer #2 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2513.7 | Semi standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #2 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2647.8 | Standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #2 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4198.9 | Standard polar | 33892256 |
| Sepiapterin,4TMS,isomer #3 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2436.4 | Semi standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #3 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2616.1 | Standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #3 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 4078.6 | Standard polar | 33892256 |
| Sepiapterin,4TMS,isomer #4 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O | 2384.8 | Semi standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #4 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O | 2580.0 | Standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #4 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O | 3728.2 | Standard polar | 33892256 |
| Sepiapterin,4TMS,isomer #5 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2436.6 | Semi standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #5 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2667.1 | Standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #5 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4052.5 | Standard polar | 33892256 |
| Sepiapterin,4TMS,isomer #6 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2365.7 | Semi standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #6 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2680.9 | Standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #6 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 3865.1 | Standard polar | 33892256 |
| Sepiapterin,4TMS,isomer #7 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2387.9 | Semi standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #7 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2664.1 | Standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #7 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 3946.4 | Standard polar | 33892256 |
| Sepiapterin,4TMS,isomer #8 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2489.0 | Semi standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #8 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2668.3 | Standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #8 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4033.5 | Standard polar | 33892256 |
| Sepiapterin,4TMS,isomer #9 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2432.9 | Semi standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #9 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2649.9 | Standard non polar | 33892256 |
| Sepiapterin,4TMS,isomer #9 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 3876.8 | Standard polar | 33892256 |
| Sepiapterin,5TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2542.2 | Semi standard non polar | 33892256 |
| Sepiapterin,5TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2684.5 | Standard non polar | 33892256 |
| Sepiapterin,5TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 3825.5 | Standard polar | 33892256 |
| Sepiapterin,5TMS,isomer #2 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2480.6 | Semi standard non polar | 33892256 |
| Sepiapterin,5TMS,isomer #2 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 2674.2 | Standard non polar | 33892256 |
| Sepiapterin,5TMS,isomer #2 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N[Si](C)(C)C)=NC2=O | 3658.5 | Standard polar | 33892256 |
| Sepiapterin,5TMS,isomer #3 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2505.9 | Semi standard non polar | 33892256 |
| Sepiapterin,5TMS,isomer #3 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 2712.9 | Standard non polar | 33892256 |
| Sepiapterin,5TMS,isomer #3 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1 | 3745.9 | Standard polar | 33892256 |
| Sepiapterin,5TMS,isomer #4 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2467.2 | Semi standard non polar | 33892256 |
| Sepiapterin,5TMS,isomer #4 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2744.8 | Standard non polar | 33892256 |
| Sepiapterin,5TMS,isomer #4 | C[C@H](O[Si](C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 3525.4 | Standard polar | 33892256 |
| Sepiapterin,5TMS,isomer #5 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2509.1 | Semi standard non polar | 33892256 |
| Sepiapterin,5TMS,isomer #5 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2756.1 | Standard non polar | 33892256 |
| Sepiapterin,5TMS,isomer #5 | CC(O)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 3520.5 | Standard polar | 33892256 |
| Sepiapterin,6TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2580.0 | Semi standard non polar | 33892256 |
| Sepiapterin,6TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 2798.3 | Standard non polar | 33892256 |
| Sepiapterin,6TMS,isomer #1 | CC(O[Si](C)(C)C)=C(O[Si](C)(C)C)C1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 3365.4 | Standard polar | 33892256 |
| Sepiapterin,1TBDMS,isomer #1 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N)=NC2=O | 2635.6 | Semi standard non polar | 33892256 |
| Sepiapterin,1TBDMS,isomer #2 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N)=NC2=O | 2692.7 | Semi standard non polar | 33892256 |
| Sepiapterin,1TBDMS,isomer #3 | C[C@H](O)C(=O)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O | 2643.9 | Semi standard non polar | 33892256 |
| Sepiapterin,1TBDMS,isomer #4 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 2523.8 | Semi standard non polar | 33892256 |
| Sepiapterin,1TBDMS,isomer #5 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 2623.1 | Semi standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #1 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N)=NC2=O | 2869.2 | Semi standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #1 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N)=NC2=O | 2868.5 | Standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #1 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N)=NC2=O | 4195.8 | Standard polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #10 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 2693.8 | Semi standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #10 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3010.3 | Standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #10 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 4523.3 | Standard polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #11 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 2694.0 | Semi standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #11 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 2883.6 | Standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #11 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 4067.2 | Standard polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #2 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O | 2779.8 | Semi standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #2 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3047.3 | Standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #2 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O | 4695.3 | Standard polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #3 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 2804.2 | Semi standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #3 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 2893.1 | Standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #3 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 4191.5 | Standard polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #4 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 2674.6 | Semi standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #4 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 2930.8 | Standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #4 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 4081.5 | Standard polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #5 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O | 2864.9 | Semi standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #5 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O | 2977.8 | Standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #5 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O | 4731.0 | Standard polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #6 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 2850.9 | Semi standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #6 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 2870.2 | Standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #6 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 4215.1 | Standard polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #7 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 2749.9 | Semi standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #7 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 2905.5 | Standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #7 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 4105.8 | Standard polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #8 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 2807.6 | Semi standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #8 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 2990.2 | Standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #8 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 4485.6 | Standard polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #9 | C[C@H](O)C(=O)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 2784.7 | Semi standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #9 | C[C@H](O)C(=O)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3005.7 | Standard non polar | 33892256 |
| Sepiapterin,2TBDMS,isomer #9 | C[C@H](O)C(=O)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 4643.4 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #1 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3023.5 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #1 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3150.3 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #1 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O | 4514.7 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #10 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 2963.3 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #10 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3192.9 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #10 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 4329.3 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #11 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 2983.1 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #11 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 3111.4 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #11 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 3923.2 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #12 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 2985.6 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #12 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3221.2 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #12 | C[C@H](O)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 4154.7 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #13 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 2911.3 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #13 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3243.1 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #13 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 4079.5 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #14 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 2858.4 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #14 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3195.9 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #14 | C[C@H](O)C(=O)C1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 4178.3 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #2 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 3058.6 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #2 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 3051.7 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #2 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 4022.9 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #3 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 2894.5 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #3 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3082.8 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #3 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3907.1 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #4 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 2987.2 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #4 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3227.2 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #4 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 4276.1 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #5 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 2927.2 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #5 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3267.7 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #5 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 4385.0 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #6 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C([NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 2871.3 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #6 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C([NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3230.2 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #6 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C([NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 4312.8 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #7 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 2915.4 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #7 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 3124.3 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #7 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 3901.9 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #8 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3069.1 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #8 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3146.4 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #8 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 4282.4 | Standard polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #9 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3009.5 | Semi standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #9 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3200.9 | Standard non polar | 33892256 |
| Sepiapterin,3TBDMS,isomer #9 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 4379.1 | Standard polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #1 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3251.9 | Semi standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #1 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3266.0 | Standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #1 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 4110.3 | Standard polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #10 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3134.4 | Semi standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #10 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3401.5 | Standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #10 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3995.6 | Standard polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #11 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3122.0 | Semi standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #11 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3412.1 | Standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #11 | C[C@H](O)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3792.1 | Standard polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #2 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3168.3 | Semi standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #2 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3362.1 | Standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #2 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 4185.9 | Standard polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #3 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3128.9 | Semi standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #3 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3354.6 | Standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #3 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 4145.0 | Standard polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #4 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 3189.4 | Semi standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #4 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 3275.8 | Standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #4 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 3840.2 | Standard polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #5 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3173.2 | Semi standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #5 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3417.6 | Standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #5 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3992.1 | Standard polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #6 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3124.9 | Semi standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #6 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3415.4 | Standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #6 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3909.0 | Standard polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #7 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3070.2 | Semi standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #7 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3403.9 | Standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #7 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3995.8 | Standard polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #8 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3237.8 | Semi standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #8 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3343.6 | Standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #8 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3987.1 | Standard polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #9 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3195.2 | Semi standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #9 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3363.6 | Standard non polar | 33892256 |
| Sepiapterin,4TBDMS,isomer #9 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3917.7 | Standard polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #1 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3422.1 | Semi standard non polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #1 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3478.8 | Standard non polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #1 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3889.7 | Standard polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #2 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3373.7 | Semi standard non polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #2 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3506.5 | Standard non polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #2 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N[Si](C)(C)C(C)(C)C)=NC2=O | 3827.5 | Standard polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #3 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3321.8 | Semi standard non polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #3 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3583.9 | Standard non polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #3 | CC(O[Si](C)(C)C(C)(C)C)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1 | 3889.3 | Standard polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #4 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3341.8 | Semi standard non polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #4 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3597.1 | Standard non polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #4 | C[C@H](O[Si](C)(C)C(C)(C)C)C(=O)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3703.6 | Standard polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #5 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3389.8 | Semi standard non polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #5 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3591.5 | Standard non polar | 33892256 |
| Sepiapterin,5TBDMS,isomer #5 | CC(O)=C(O[Si](C)(C)C(C)(C)C)C1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 3714.4 | Standard polar | 33892256 |