| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:49:05 UTC |
|---|
| HMDB ID | HMDB0000673 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Linoleic acid |
|---|
| Description | Linoleic acid is a doubly unsaturated fatty acid, also known as an omega-6 fatty acid, occurring widely in plant glycosides. In this particular polyunsaturated fatty acid (PUFA), the first double bond is located between the sixth and seventh carbon atom from the methyl end of the fatty acid (n-6). Linoleic acid is an essential fatty acid in human nutrition because it cannot be synthesized by humans. It is used in the biosynthesis of prostaglandins (via arachidonic acid) and cell membranes (From Stedman, 26th ed). Linoleic acid is found to be associated with isovaleric acidemia, which is an inborn error of metabolism. |
|---|
| Structure | CCCCC\C=C/C\C=C/CCCCCCCC(O)=O InChI=1S/C18H32O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18(19)20/h6-7,9-10H,2-5,8,11-17H2,1H3,(H,19,20)/b7-6-,10-9- |
|---|
| Synonyms | | Value | Source |
|---|
| (9Z,12Z)-Octadecadienoic acid | ChEBI | | (Z,Z)-9,12-Octadecadienoic acid | ChEBI | | 9-cis,12-cis-Octadecadienoic acid | ChEBI | | 9Z,12Z-Octadecadienoic acid | ChEBI | | Acide cis-linoleique | ChEBI | | Acide linoleique | ChEBI | | Acido linoleico | ChEBI | | all-cis-9,12-Octadecadienoic acid | ChEBI | | C18:2 9C, 12C Omega6 todos cis-9,12-octadienoico | ChEBI | | C18:2, N-6,9 all-cis | ChEBI | | cis,cis-9,12-Octadecadienoic acid | ChEBI | | cis,cis-Linoleic acid | ChEBI | | cis-Delta(9,12)-Octadecadienoic acid | ChEBI | | LA | ChEBI | | Linolic acid | ChEBI | | 9-cis,12-cis-Octadecadienoate | Kegg | | (9Z,12Z)-Octadecadienoate | Generator | | (Z,Z)-9,12-Octadecadienoate | Generator | | 9Z,12Z-Octadecadienoate | Generator | | all-cis-9,12-Octadecadienoate | Generator | | cis,cis-9,12-Octadecadienoate | Generator | | cis,cis-Linoleate | Generator | | cis-delta(9,12)-Octadecadienoate | Generator | | cis-Δ(9,12)-octadecadienoate | Generator | | cis-Δ(9,12)-octadecadienoic acid | Generator | | Linolate | Generator | | Linoleate | Generator | | (9Z,12Z)-9,12-Octadecadienoate | HMDB | | (9Z,12Z)-9,12-Octadecadienoic acid | HMDB | | 9-cis,12-cis-Linoleate | HMDB | | 9-cis,12-cis-Linoleic acid | HMDB | | 9Z,12Z-Linoleate | HMDB | | 9Z,12Z-Linoleic acid | HMDB | | cis-9,cis-12-Octadecadienoate | HMDB | | cis-9,cis-12-Octadecadienoic acid | HMDB | | cis-D9,12-Octadecadienoate | HMDB | | cis-D9,12-Octadecadienoic acid | HMDB | | Emersol 315 | HMDB | | Extra linoleic 90 | HMDB | | Polylin 515 | HMDB | | Unifac 6550 | HMDB | | Acid, 9,12-octadecadienoic | HMDB | | Linoelaidic acid | HMDB | | Linoleic acid, (e,e)-isomer | HMDB | | Linoleic acid, (Z,Z)-isomer | HMDB | | Linoleic acid, (Z,Z)-isomer, 14C-labeled | HMDB | | Linoleic acid, ammonium salt, (Z,Z)-isomer | HMDB | | Linoleic acid, potassium salt, (Z,Z)-isomer | HMDB | | 9 trans,12 trans Octadecadienoic acid | HMDB | | 9-trans,12-trans-Octadecadienoic acid | HMDB | | Linoleic acid, sodium salt, (e,e)-isomer | HMDB | | Linoleic acid, sodium salt, (Z,Z)-isomer | HMDB | | trans,trans-9,12-Octadecadienoic acid | HMDB | | 9,12-Octadecadienoic acid | HMDB | | Linoelaidic acid, (e,Z)-isomer | HMDB | | Linoleic acid, calcium salt, (Z,Z)-isomer | HMDB | | Linolelaidic acid | HMDB | | 9,12 Octadecadienoic acid | HMDB | | Linoleic acid, (Z,e)-isomer | HMDB | | FA(18:2(9Z,12Z)) | HMDB |
|
|---|
| Chemical Formula | C18H32O2 |
|---|
| Average Molecular Weight | 280.4455 |
|---|
| Monoisotopic Molecular Weight | 280.240230268 |
|---|
| IUPAC Name | (9Z,12Z)-octadeca-9,12-dienoic acid |
|---|
| Traditional Name | linoleic |
|---|
| CAS Registry Number | 60-33-3 |
|---|
| SMILES | CCCCC\C=C/C\C=C/CCCCCCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C18H32O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18(19)20/h6-7,9-10H,2-5,8,11-17H2,1H3,(H,19,20)/b7-6-,10-9- |
|---|
| InChI Key | OYHQOLUKZRVURQ-HZJYTTRNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as lineolic acids and derivatives. These are derivatives of lineolic acid. Lineolic acid is a polyunsaturated omega-6 18 carbon long fatty acid, with two CC double bonds at the 9- and 12-positions. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Lineolic acids and derivatives |
|---|
| Direct Parent | Lineolic acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Octadecanoid
- Long-chain fatty acid
- Fatty acid
- Unsaturated fatty acid
- Straight chain fatty acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Not Available | Not Available |
|---|
| Physical Properties |
|---|
| State | Liquid |
|---|
| Experimental Molecular Properties | |
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized |
|---|
| Spectra |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - Linoleic acid GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (1 TMS) | splash10-000t-7900000000-b6ee03c4800464c37471 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Linoleic acid GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (1 TMS) | splash10-00vi-9300000000-c92dac639ced59eb5dbe | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Linoleic acid GC-MS (1 TMS) | splash10-003s-9700000000-77e67d7b1a161e6ecfa6 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Linoleic acid GC-EI-TOF (Non-derivatized) | splash10-000t-7900000000-b6ee03c4800464c37471 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Linoleic acid GC-EI-TOF (Non-derivatized) | splash10-00vi-9300000000-c92dac639ced59eb5dbe | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Linoleic acid GC-MS (Non-derivatized) | splash10-003s-9700000000-77e67d7b1a161e6ecfa6 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Linoleic acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-0007-9750000000-50d69948d56dd2ba6e42 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Linoleic acid GC-MS (1 TMS) - 70eV, Positive | splash10-0079-9631000000-cc93c24bbf14d81ae9b6 | 2017-10-06 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Linoleic acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Linoleic acid GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-015a-9200000000-a193c27810bedf93c498 | 2014-09-20 | Not Available | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid Quattro_QQQ 10V, Positive-QTOF (Annotated) | splash10-0f89-0190000000-2be9501b1d4a9fcbd1c0 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid Quattro_QQQ 25V, Positive-QTOF (Annotated) | splash10-0uk9-0790000000-7f6e35a591f977bc488e | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid Quattro_QQQ 40V, Positive-QTOF (Annotated) | splash10-0udi-0090000000-c56edc2bbf9d6752aec5 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid FAB-EBEB (JMS-HX/HX 110A, JEOL) , Negative-QTOF | splash10-004i-0090000000-baf4579e26c6b393d391 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative-QTOF | splash10-004i-0090000000-815c1682b2c59ba96f10 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative-QTOF | splash10-004i-0090000000-3406b2b2d5756807e1cd | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative-QTOF | splash10-004i-0090000000-2fb4975278fa4d118f43 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative-QTOF | splash10-0a6r-9380000000-37f833673c248405c8ef | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative-QTOF | splash10-0a4i-9200000000-7d34ed5900a17ffe3f9b | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid ESI-TOF 10V, Negative-QTOF | splash10-004i-0090010000-f8df6099e003402f2566 | 2017-08-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid ESI-TOF , Negative-QTOF | splash10-004i-0091021000-0e44779958d5744d873b | 2017-08-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid ESI-TOF 10V, Negative-QTOF | splash10-004i-0091021000-0e44779958d5744d873b | 2017-08-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid ESI-TOF 20V, Negative-QTOF | splash10-004i-0091021000-0e44779958d5744d873b | 2017-08-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid ESI-TOF 30V, Negative-QTOF | splash10-004i-0091021000-0e44779958d5744d873b | 2017-08-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid ESI-TOF 10V, Negative-QTOF | splash10-004i-0090010000-f8df6099e003402f2566 | 2017-09-12 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid ESI-TOF , Negative-QTOF | splash10-004i-0091021000-0e44779958d5744d873b | 2017-09-12 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid ESI-TOF 10V, Negative-QTOF | splash10-004i-0090000000-adbf36f0a17c33ac33f8 | 2017-09-12 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid ESI-TOF 20V, Negative-QTOF | splash10-004i-0090000000-2747c83af78732eb6e16 | 2017-09-12 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Linoleic acid ESI-TOF 30V, Negative-QTOF | splash10-004i-0090000000-a0415b1cb63b4562b40e | 2017-09-12 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Linoleic acid 10V, Positive-QTOF | splash10-03di-0090000000-8bdf8d54a29f73494242 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Linoleic acid 20V, Positive-QTOF | splash10-0239-4590000000-e5ee57553b064eae2efe | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Linoleic acid 40V, Positive-QTOF | splash10-00rf-9830000000-b26ba057a2b4d135b478 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Linoleic acid 10V, Negative-QTOF | splash10-004i-0090000000-f1e9e4b543f7d4f48bf8 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Linoleic acid 20V, Negative-QTOF | splash10-01ti-0090000000-665523c6142ff4e39c96 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Linoleic acid 40V, Negative-QTOF | splash10-0a4l-9240000000-dca36a25ad7519d500c3 | 2017-09-01 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CDCl3, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
|
|---|
| Biological Properties |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Membrane (predicted from logP)
|
|---|
| Biospecimen Locations | - Blood
- Cerebrospinal Fluid (CSF)
- Feces
- Saliva
- Urine
|
|---|
| Tissue Locations | - Adipose Tissue
- Epidermis
- Erythrocyte
- Fibroblasts
- Intestine
- Kidney
- Neuron
- Placenta
- Platelet
- Prostate
- Skeletal Muscle
- Spleen
|
|---|
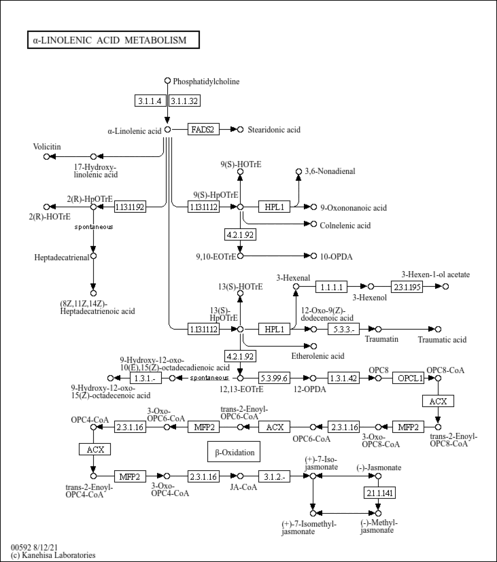
| Pathways | |
|---|
| Normal Concentrations |
|---|
| |
| Blood | Detected and Quantified | 23.39 +/- 2.75 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 110 (42-370) uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 83.8 +/- 38 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 193.7 +/- 72.0 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 264 +/- 17.0 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 183.7 +/- 69.9 uM | Adult (>18 years old) | Male | Normal | | details | | Blood | Detected and Quantified | 207.9 +/- 73.0 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 734.546 +/- 92.710 uM | Adult (>18 years old) | Male | Normal | | details | | Blood | Detected and Quantified | 798.729 +/- 99.841 uM | Adult (>18 years old) | Male | Normal | | details | | Blood | Detected and Quantified | 927.0964 +/- 110.538 uM | Adult (>18 years old) | Male | Normal | | details | | Blood | Detected and Quantified | 930.662 +/- 89.144 uM | Adult (>18 years old) | Male | Normal | | details | | Blood | Detected and Quantified | 61 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 17.7 +/- 4.7 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 14.733 +/- 4.331 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 74 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 134 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 192 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 246 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 291 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 308 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 311 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 11.8-21.6 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 525 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 563 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 618 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 1021.482 +/- 224.215 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 1065.519 +/- 201.893 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 15.2 +/- 0.437 uM | Adult (>18 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 11.0 +/- 3.0 uM | Adult (>18 years old) | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Infant (0-1 year old) | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Infant (0-1 year old) | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Children (1-13 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Not Specified | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Infant (0-1 year old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Not Specified | Not Specified | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Not Specified | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 0.05 +/- 0.05 umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 1.19 +/- 0.61 umol/mmol creatinine | Adult (>18 years old) | Female | Normal | | details |
|
|---|
| Abnormal Concentrations |
|---|
| |
| Blood | Detected and Quantified | 22.61 +/- 2.80 uM | Adult (>18 years old) | Female | Gestational diabetes mellitus (GDM) | | details | | Blood | Detected and Quantified | 886.0 +/- 626.6 uM | Adult (>18 years old) | Both | Cirrhosis | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Oesophageal cancer | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Lung cancer | | details | | Blood | Detected and Quantified | 175.9 +/- 65.0 uM | Adult (>18 years old) | Both | Hypertension | | details | | Blood | Detected and Quantified | 159 +/- 12.0 uM | Adult (>18 years old) | Both | Schizophrenia | | details | | Blood | Detected and Quantified | 2796.7 +/- 493.9 uM | Adult (>18 years old) | Not Specified | Isovaleric acidemia | | details | | Blood | Detected and Quantified | 167.0 +/- 63.8 uM | Adult (>18 years old) | Male | Essential hypertension | | details | | Blood | Detected and Quantified | 195.2 +/- 64.4 uM | Adult (>18 years old) | Female | Essential hypertension | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Schizophrenia | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Female | Breast cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | CCD | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Ileal Crohn's disease | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | liver cirrhosis | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal Cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Newborn (0-30 days old) | Not Specified | Premature neonates | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Ulcerative colitis | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Female | Interstitial cystitis | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Urine | Detected and Quantified | 0.18 +/- 0.45 umol/mmol creatinine | Adult (>18 years old) | Female | Thyroid cancer | | details |
|
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | | Gestational diabetes |
|---|
- Min Y, Ghebremeskel K, Lowy C, Thomas B, Crawford MA: Adverse effect of obesity on red cell membrane arachidonic and docosahexaenoic acids in gestational diabetes. Diabetologia. 2004 Jan;47(1):75-81. Epub 2003 Nov 22. [PubMed:14634727 ]
| | Cirrhosis |
|---|
- Szebeni J, Eskelson C, Sampliner R, Hartmann B, Griffin J, Dormandy T, Watson RR: Plasma fatty acid pattern including diene-conjugated linoleic acid in ethanol users and patients with ethanol-related liver disease. Alcohol Clin Exp Res. 1986 Dec;10(6):647-50. [PubMed:3101533 ]
- Huang HJ, Zhang AY, Cao HC, Lu HF, Wang BH, Xie Q, Xu W, Li LJ: Metabolomic analyses of faeces reveals malabsorption in cirrhotic patients. Dig Liver Dis. 2013 Aug;45(8):677-82. doi: 10.1016/j.dld.2013.01.001. Epub 2013 Feb 4. [PubMed:23384618 ]
| | Hypertension |
|---|
- Wang S, Ma A, Song S, Quan Q, Zhao X, Zheng X: Fasting serum free fatty acid composition, waist/hip ratio and insulin activity in essential hypertensive patients. Hypertens Res. 2008 Apr;31(4):623-32. doi: 10.1291/hypres.31.623. [PubMed:18633173 ]
| | Essential hypertension |
|---|
- Wang S, Ma A, Song S, Quan Q, Zhao X, Zheng X: Fasting serum free fatty acid composition, waist/hip ratio and insulin activity in essential hypertensive patients. Hypertens Res. 2008 Apr;31(4):623-32. doi: 10.1291/hypres.31.623. [PubMed:18633173 ]
| | Perillyl alcohol administration for cancer treatment |
|---|
- Lv W, Yang T: Identification of possible biomarkers for breast cancer from free fatty acid profiles determined by GC-MS and multivariate statistical analysis. Clin Biochem. 2012 Jan;45(1-2):127-33. doi: 10.1016/j.clinbiochem.2011.10.011. Epub 2011 Oct 26. [PubMed:22061338 ]
| | Lung Cancer |
|---|
- Chen Y, Ma Z, Min L, Li H, Wang B, Zhong J, Dai L: Biomarker identification and pathway analysis by serum metabolomics of lung cancer. Biomed Res Int. 2015;2015:183624. doi: 10.1155/2015/183624. Epub 2015 Apr 16. [PubMed:25961003 ]
| | Schizophrenia |
|---|
- Fukushima T, Iizuka H, Yokota A, Suzuki T, Ohno C, Kono Y, Nishikiori M, Seki A, Ichiba H, Watanabe Y, Hongo S, Utsunomiya M, Nakatani M, Sadamoto K, Yoshio T: Quantitative analyses of schizophrenia-associated metabolites in serum: serum D-lactate levels are negatively correlated with gamma-glutamylcysteine in medicated schizophrenia patients. PLoS One. 2014 Jul 8;9(7):e101652. doi: 10.1371/journal.pone.0101652. eCollection 2014. [PubMed:25004141 ]
- Xuan J, Pan G, Qiu Y, Yang L, Su M, Liu Y, Chen J, Feng G, Fang Y, Jia W, Xing Q, He L: Metabolomic profiling to identify potential serum biomarkers for schizophrenia and risperidone action. J Proteome Res. 2011 Dec 2;10(12):5433-43. doi: 10.1021/pr2006796. Epub 2011 Nov 8. [PubMed:22007635 ]
| | Isovaleric acidemia |
|---|
- Dercksen M, Kulik W, Mienie LJ, Reinecke CJ, Wanders RJ, Duran M: Polyunsaturated fatty acid status in treated isovaleric acidemia patients. Eur J Clin Nutr. 2016 Oct;70(10):1123-1126. doi: 10.1038/ejcn.2016.100. Epub 2016 Jun 22. [PubMed:27329611 ]
| | Colorectal cancer |
|---|
- Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP: Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013 Aug 6;8(8):e70803. doi: 10.1371/journal.pone.0070803. Print 2013. [PubMed:23940645 ]
- Phua LC, Chue XP, Koh PK, Cheah PY, Ho HK, Chan EC: Non-invasive fecal metabonomic detection of colorectal cancer. Cancer Biol Ther. 2014 Apr;15(4):389-97. doi: 10.4161/cbt.27625. Epub 2014 Jan 14. [PubMed:24424155 ]
- Ni Y, Xie G, Jia W: Metabonomics of human colorectal cancer: new approaches for early diagnosis and biomarker discovery. J Proteome Res. 2014 Sep 5;13(9):3857-70. doi: 10.1021/pr500443c. Epub 2014 Aug 14. [PubMed:25105552 ]
- Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP: Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016 Jun 6;4:11. doi: 10.1186/s40170-016-0151-y. eCollection 2016. [PubMed:27275383 ]
- Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ: Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016 Mar 25;11(3):e0152126. doi: 10.1371/journal.pone.0152126. eCollection 2016. [PubMed:27015276 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
- Wang X, Wang J, Rao B, Deng L: Gut flora profiling and fecal metabolite composition of colorectal cancer patients and healthy individuals. Exp Ther Med. 2017 Jun;13(6):2848-2854. doi: 10.3892/etm.2017.4367. Epub 2017 Apr 20. [PubMed:28587349 ]
| | Bladder infections |
|---|
- Braundmeier-Fleming A, Russell NT, Yang W, Nas MY, Yaggie RE, Berry M, Bachrach L, Flury SC, Marko DS, Bushell CB, Welge ME, White BA, Schaeffer AJ, Klumpp DJ: Stool-based biomarkers of interstitial cystitis/bladder pain syndrome. Sci Rep. 2016 May 18;6:26083. doi: 10.1038/srep26083. [PubMed:27188581 ]
| | Ulcerative colitis |
|---|
- Azario I, Pievani A, Del Priore F, Antolini L, Santi L, Corsi A, Cardinale L, Sawamoto K, Kubaski F, Gentner B, Bernardo ME, Valsecchi MG, Riminucci M, Tomatsu S, Aiuti A, Biondi A, Serafini M: Neonatal umbilical cord blood transplantation halts skeletal disease progression in the murine model of MPS-I. Sci Rep. 2017 Aug 25;7(1):9473. doi: 10.1038/s41598-017-09958-9. [PubMed:28842642 ]
| | Thyroid cancer |
|---|
- Kim KM, Jung BH, Lho DS, Chung WY, Paeng KJ, Chung BC: Alteration of urinary profiles of endogenous steroids and polyunsaturated fatty acids in thyroid cancer. Cancer Lett. 2003 Dec 30;202(2):173-9. [PubMed:14643447 ]
|
|
|---|
| Associated OMIM IDs | |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB006287 |
|---|
| KNApSAcK ID | C00001224 |
|---|
| Chemspider ID | 4444105 |
|---|
| KEGG Compound ID | C01595 |
|---|
| BioCyc ID | LINOLEIC_ACID |
|---|
| BiGG ID | 37956 |
|---|
| Wikipedia Link | Linoleic_acid |
|---|
| METLIN ID | 191 |
|---|
| PubChem Compound | 5280450 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17351 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | LNLC |
|---|
| MarkerDB ID | MDB00000213 |
|---|
| Good Scents ID | rw1045901 |
|---|
| References |
|---|
| Synthesis Reference | Walborsky, Harry M.; Davis, Robert H.; Howton, David R. A total synthesis of linoleic acid. Journal of the American Chemical Society (1951), 73 2590-4. |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| General References | - Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. doi: 10.1038/nature07762. [PubMed:19212411 ]
- Hoffmann GF, Meier-Augenstein W, Stockler S, Surtees R, Rating D, Nyhan WL: Physiology and pathophysiology of organic acids in cerebrospinal fluid. J Inherit Metab Dis. 1993;16(4):648-69. [PubMed:8412012 ]
- Schurer NY, Stremmel W, Grundmann JU, Schliep V, Kleinert H, Bass NM, Williams ML: Evidence for a novel keratinocyte fatty acid uptake mechanism with preference for linoleic acid: comparison of oleic and linoleic acid uptake by cultured human keratinocytes, fibroblasts and a human hepatoma cell line. Biochim Biophys Acta. 1994 Feb 10;1211(1):51-60. [PubMed:8123682 ]
- Valianpour F, Wanders RJ, Overmars H, Vaz FM, Barth PG, van Gennip AH: Linoleic acid supplementation of Barth syndrome fibroblasts restores cardiolipin levels: implications for treatment. J Lipid Res. 2003 Mar;44(3):560-6. Epub 2002 Dec 16. [PubMed:12562862 ]
- Horrobin DF: Essential fatty acid metabolism and its modification in atopic eczema. Am J Clin Nutr. 2000 Jan;71(1 Suppl):367S-72S. [PubMed:10617999 ]
- Imokawa G, Yada Y, Higuchi K, Okuda M, Ohashi Y, Kawamata A: Pseudo-acylceramide with linoleic acid produces selective recovery of diminished cutaneous barrier function in essential fatty acid-deficient rats and has an inhibitory effect on epidermal hyperplasia. J Clin Invest. 1994 Jul;94(1):89-96. [PubMed:8040295 ]
- Kawajiri H, Hsi LC, Kamitani H, Ikawa H, Geller M, Ward T, Eling TE, Glasgow WC: Arachidonic and linoleic acid metabolism in mouse intestinal tissue: evidence for novel lipoxygenase activity. Arch Biochem Biophys. 2002 Feb 1;398(1):51-60. [PubMed:11811948 ]
- Iso H, Sato S, Umemura U, Kudo M, Koike K, Kitamura A, Imano H, Okamura T, Naito Y, Shimamoto T: Linoleic acid, other fatty acids, and the risk of stroke. Stroke. 2002 Aug;33(8):2086-93. [PubMed:12154268 ]
- Seidel D, Heipertz R, Weisner B: Cerebrospinal fluid lipids in demyelinating disease. II. Linoleic acid as an index of impaired blood-CSF barrier. J Neurol. 1980 Jan;222(3):177-82. [PubMed:6153705 ]
- Salo P, Seppanen-Laakso T, Laakso I, Seppanen R, Niinikoski H, Viikari J, Simell O: Low-saturated fat, low-cholesterol diet in 3-year-old children: effect on intake and composition of trans fatty acids and other fatty acids in serum phospholipid fraction-The STRIP study. Special Turku coronary Risk factor Intervention Project for children. J Pediatr. 2000 Jan;136(1):46-52. [PubMed:10636973 ]
- Grimsgaard S, Bonaa KH, Jacobsen BK, Bjerve KS: Plasma saturated and linoleic fatty acids are independently associated with blood pressure. Hypertension. 1999 Sep;34(3):478-83. [PubMed:10489397 ]
- Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. [PubMed:32033212 ]
|
|---|