| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2008-10-16 22:22:16 UTC |
|---|
| Update Date | 2023-02-21 17:17:26 UTC |
|---|
| HMDB ID | HMDB0011103 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 1,7-Dimethyluric acid |
|---|
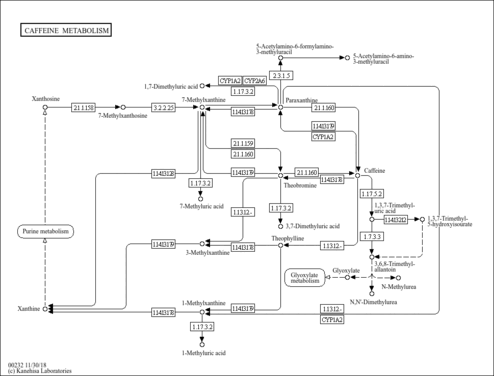
| Description | 1,7-Dimethyluric acid, also known as 1,7-dimethylate, belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. 1,7-Dimethyluric acid is an extremely weak basic (essentially neutral) compound (based on its pKa). 1,7-Dimethyluric acid exists in all living organisms, ranging from bacteria to humans. 1,7-dimethyluric acid can be biosynthesized from paraxanthine through the action of the enzymes cytochrome P450 1A2 and cytochrome P450 2A6. In humans, 1,7-dimethyluric acid is involved in caffeine metabolism. |
|---|
| Structure | CN1C(=O)NC2=C1C(=O)N(C)C(=O)N2 InChI=1S/C7H8N4O3/c1-10-3-4(8-6(10)13)9-7(14)11(2)5(3)12/h1-2H3,(H,8,13)(H,9,14) |
|---|
| Synonyms | | Value | Source |
|---|
| 1,7-Dimethylate | Generator | | 1,7-Dimethylic acid | Generator | | 1,7-Dimethylurate | HMDB | | 7,9-Dihydro-1,7-dimethyl-1H-purine-2,6,8(3H)-trione | HMDB | | 1,7-Dimethyluric acid | HMDB |
|
|---|
| Chemical Formula | C7H8N4O3 |
|---|
| Average Molecular Weight | 196.1634 |
|---|
| Monoisotopic Molecular Weight | 196.059640142 |
|---|
| IUPAC Name | 2,8-dihydroxy-1,7-dimethyl-6,7-dihydro-1H-purin-6-one |
|---|
| Traditional Name | 1,7-dimethyluric acid |
|---|
| CAS Registry Number | 33868-03-0 |
|---|
| SMILES | CN1C(=O)NC2=C1C(=O)N(C)C(=O)N2 |
|---|
| InChI Identifier | InChI=1S/C7H8N4O3/c1-10-3-4(8-6(10)13)9-7(14)11(2)5(3)12/h1-2H3,(H,8,13)(H,9,14) |
|---|
| InChI Key | NOFNCLGCUJJPKU-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Xanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthine
- 6-oxopurine
- Purinone
- Alkaloid or derivatives
- Pyrimidone
- Hydroxypyrimidine
- N-substituted imidazole
- Pyrimidine
- Imidazole
- Azole
- Heteroaromatic compound
- Lactam
- Azacycle
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. | 2.38 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 9.2084 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 3.07 minutes | 32390414 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 659.5 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 288.0 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 74.7 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 165.4 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 46.9 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 250.3 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 295.3 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 80.7 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 611.4 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 290.5 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 784.2 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 206.5 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 180.4 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 596.2 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 184.7 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 156.4 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| 1,7-Dimethyluric acid,1TMS,isomer #1 | CN1C(=O)[NH]C2=C(C1=O)N(C)C(=O)N2[Si](C)(C)C | 1999.7 | Semi standard non polar | 33892256 | | 1,7-Dimethyluric acid,1TMS,isomer #1 | CN1C(=O)[NH]C2=C(C1=O)N(C)C(=O)N2[Si](C)(C)C | 2240.9 | Standard non polar | 33892256 | | 1,7-Dimethyluric acid,1TMS,isomer #1 | CN1C(=O)[NH]C2=C(C1=O)N(C)C(=O)N2[Si](C)(C)C | 2881.8 | Standard polar | 33892256 | | 1,7-Dimethyluric acid,1TMS,isomer #2 | CN1C(=O)C2=C([NH]C(=O)N2C)N([Si](C)(C)C)C1=O | 1985.9 | Semi standard non polar | 33892256 | | 1,7-Dimethyluric acid,1TMS,isomer #2 | CN1C(=O)C2=C([NH]C(=O)N2C)N([Si](C)(C)C)C1=O | 2248.6 | Standard non polar | 33892256 | | 1,7-Dimethyluric acid,1TMS,isomer #2 | CN1C(=O)C2=C([NH]C(=O)N2C)N([Si](C)(C)C)C1=O | 2859.3 | Standard polar | 33892256 | | 1,7-Dimethyluric acid,2TMS,isomer #1 | CN1C(=O)C2=C(N([Si](C)(C)C)C1=O)N([Si](C)(C)C)C(=O)N2C | 1985.9 | Semi standard non polar | 33892256 | | 1,7-Dimethyluric acid,2TMS,isomer #1 | CN1C(=O)C2=C(N([Si](C)(C)C)C1=O)N([Si](C)(C)C)C(=O)N2C | 2286.7 | Standard non polar | 33892256 | | 1,7-Dimethyluric acid,2TMS,isomer #1 | CN1C(=O)C2=C(N([Si](C)(C)C)C1=O)N([Si](C)(C)C)C(=O)N2C | 2470.8 | Standard polar | 33892256 | | 1,7-Dimethyluric acid,1TBDMS,isomer #1 | CN1C(=O)[NH]C2=C(C1=O)N(C)C(=O)N2[Si](C)(C)C(C)(C)C | 2205.1 | Semi standard non polar | 33892256 | | 1,7-Dimethyluric acid,1TBDMS,isomer #1 | CN1C(=O)[NH]C2=C(C1=O)N(C)C(=O)N2[Si](C)(C)C(C)(C)C | 2449.9 | Standard non polar | 33892256 | | 1,7-Dimethyluric acid,1TBDMS,isomer #1 | CN1C(=O)[NH]C2=C(C1=O)N(C)C(=O)N2[Si](C)(C)C(C)(C)C | 2829.0 | Standard polar | 33892256 | | 1,7-Dimethyluric acid,1TBDMS,isomer #2 | CN1C(=O)C2=C([NH]C(=O)N2C)N([Si](C)(C)C(C)(C)C)C1=O | 2203.7 | Semi standard non polar | 33892256 | | 1,7-Dimethyluric acid,1TBDMS,isomer #2 | CN1C(=O)C2=C([NH]C(=O)N2C)N([Si](C)(C)C(C)(C)C)C1=O | 2454.8 | Standard non polar | 33892256 | | 1,7-Dimethyluric acid,1TBDMS,isomer #2 | CN1C(=O)C2=C([NH]C(=O)N2C)N([Si](C)(C)C(C)(C)C)C1=O | 2810.5 | Standard polar | 33892256 | | 1,7-Dimethyluric acid,2TBDMS,isomer #1 | CN1C(=O)C2=C(N([Si](C)(C)C(C)(C)C)C1=O)N([Si](C)(C)C(C)(C)C)C(=O)N2C | 2406.2 | Semi standard non polar | 33892256 | | 1,7-Dimethyluric acid,2TBDMS,isomer #1 | CN1C(=O)C2=C(N([Si](C)(C)C(C)(C)C)C1=O)N([Si](C)(C)C(C)(C)C)C(=O)N2C | 2714.1 | Standard non polar | 33892256 | | 1,7-Dimethyluric acid,2TBDMS,isomer #1 | CN1C(=O)C2=C(N([Si](C)(C)C(C)(C)C)C1=O)N([Si](C)(C)C(C)(C)C)C(=O)N2C | 2578.0 | Standard polar | 33892256 |
|
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - 1,7-Dimethyluric acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-029l-0900000000-fa683b3e4ecb7c70f206 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 1,7-Dimethyluric acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 1,7-Dimethyluric acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid LC-ESI-QFT , positive-QTOF | splash10-0002-0900000000-f40482c57d127e32f8e4 | 2020-07-21 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid LC-ESI-QTOF 35V, positive-QTOF | splash10-0007-0900000000-6860551c1745ba41864f | 2020-07-21 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid NA , positive-QTOF | splash10-0002-0900000000-c180cf8aed0912f88a75 | 2020-07-21 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid LC-ESI-QFT , negative-QTOF | splash10-0002-0900000000-ae0b921d4ce14cd8443d | 2020-07-21 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid LC-ESI-QTOF 35V, negative-QTOF | splash10-001i-0900000000-9d422231f3b705d6e815 | 2020-07-21 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid Orbitrap 4V, positive-QTOF | splash10-0002-0900000000-440ffa8e3cca49f55cc5 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid Orbitrap 5V, positive-QTOF | splash10-0002-0900000000-9191d6a6a1abdd23d811 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid Orbitrap 5V, positive-QTOF | splash10-0002-0900000000-1b5e888daf17a0165e55 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid Orbitrap 6V, positive-QTOF | splash10-0005-0900000000-df2cb96313fe3610cc44 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid Orbitrap 7V, positive-QTOF | splash10-0007-0900000000-6234b012018c1ae4d86e | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid Orbitrap 9V, positive-QTOF | splash10-0006-1900000000-86334014f96303c72304 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid Orbitrap 11V, positive-QTOF | splash10-0006-6900000000-ca9b5373df8e0ea8fcd3 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid Orbitrap 13V, positive-QTOF | splash10-01bc-9300000000-7a850dc063e0bf5fd5fb | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid n/a 13V, positive-QTOF | splash10-0006-0900000000-0277172e5e710f4775de | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid n/a 13V, positive-QTOF | splash10-03di-6900000000-542cf592e0459e911f5c | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid n/a 13V, positive-QTOF | splash10-00xr-9000000000-db67735b192b87b601b3 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid n/a 13V, positive-QTOF | splash10-000i-0900000000-e0b8225c7958cf0367cf | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid n/a 13V, positive-QTOF | splash10-03di-0900000000-8cd35e2b2191f4fb94a1 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,7-Dimethyluric acid n/a 13V, positive-QTOF | splash10-004s-3900000000-f3c613d69643e20758ce | 2020-07-22 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1,7-Dimethyluric acid 10V, Positive-QTOF | splash10-0002-0900000000-bd55edf6a44d0c64f3b1 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1,7-Dimethyluric acid 20V, Positive-QTOF | splash10-0002-0900000000-1ba40353172a8d8a0d53 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1,7-Dimethyluric acid 40V, Positive-QTOF | splash10-0002-9600000000-9845b1cfa11ec4dcced0 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1,7-Dimethyluric acid 10V, Negative-QTOF | splash10-0002-1900000000-45f918e65bf50b9c83ce | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1,7-Dimethyluric acid 20V, Negative-QTOF | splash10-0002-2900000000-683f1d7f14c8b26bd2cd | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1,7-Dimethyluric acid 40V, Negative-QTOF | splash10-05mp-9200000000-9022272eeb07eb313dfd | 2016-08-03 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
|
|---|