| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:49:09 UTC |
|---|
| HMDB ID | HMDB0001275 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Propionyl-CoA |
|---|
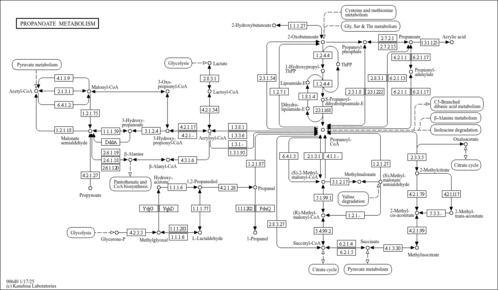
| Description | Propionyl-CoA is an intermediate in the metabolism of propanoate. Propionic aciduria is caused by an autosomal recessive disorder of propionyl coenzyme A (CoA) carboxylase deficiency (EC 6.4.1.3). In propionic aciduria, propionyl CoA accumulates within the mitochondria in massive quantities; free carnitine is then esterified, creating propionyl carnitine, which is then excreted in the urine. Because the supply of carnitine in the diet and from synthesis is limited, such patients readily develop carnitine deficiency as a result of the increased loss of acylcarnitine derivatives. This condition demands supplementation of free carnitine above the normal dietary intake to continue to remove (detoxify) the accumulating organic acids. Propionyl-CoA is a substrate for Acyl-CoA dehydrogenase (medium-chain specific, mitochondrial), Acetyl-coenzyme A synthetase 2-like (mitochondrial), Propionyl-CoA carboxylase alpha chain (mitochondrial), Methylmalonate-semialdehyde dehydrogenase (mitochondrial), Trifunctional enzyme beta subunit (mitochondrial), 3-ketoacyl-CoA thiolase (peroxisomal), Acyl-CoA dehydrogenase (long-chain specific, mitochondrial), Malonyl-CoA decarboxylase (mitochondrial), Acetyl-coenzyme A synthetase (cytoplasmic), 3-ketoacyl-CoA thiolase (mitochondrial) and Propionyl-CoA carboxylase beta chain (mitochondrial). (PMID: 10650319 ). |
|---|
| Structure | CCC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N InChI=1S/C24H40N7O17P3S/c1-4-15(33)52-8-7-26-14(32)5-6-27-22(36)19(35)24(2,3)10-45-51(42,43)48-50(40,41)44-9-13-18(47-49(37,38)39)17(34)23(46-13)31-12-30-16-20(25)28-11-29-21(16)31/h11-13,17-19,23,34-35H,4-10H2,1-3H3,(H,26,32)(H,27,36)(H,40,41)(H,42,43)(H2,25,28,29)(H2,37,38,39)/t13-,17-,18-,19+,23-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| Propanoyl-coenzyme A | ChEBI | | Propionyl coenzyme A | ChEBI | | Propionyl-coenzyme A | ChEBI | | S-Propanoyl-CoA | ChEBI | | S-Propanoyl-coenzyme A | ChEBI | | S-Propionyl-coenzym-a | ChEBI | | S-Propionylcoenzyme A | ChEBI | | Propanoyl-CoA | ChEBI | | Propionyl CoA | HMDB | | Propionyl-CoA | HMDB | | n-Propionyl CoA | HMDB |
|
|---|
| Chemical Formula | C24H40N7O17P3S |
|---|
| Average Molecular Weight | 823.597 |
|---|
| Monoisotopic Molecular Weight | 823.141423115 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-({[hydroxy({hydroxy[(3R)-3-hydroxy-2,2-dimethyl-3-[(2-{[2-(propanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]propoxy]phosphoryl}oxy)phosphoryl]oxy}methyl)oxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | propionyl-coa |
|---|
| CAS Registry Number | 317-66-8 |
|---|
| SMILES | CCC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C24H40N7O17P3S/c1-4-15(33)52-8-7-26-14(32)5-6-27-22(36)19(35)24(2,3)10-45-51(42,43)48-50(40,41)44-9-13-18(47-49(37,38)39)17(34)23(46-13)31-12-30-16-20(25)28-11-29-21(16)31/h11-13,17-19,23,34-35H,4-10H2,1-3H3,(H,26,32)(H,27,36)(H,40,41)(H,42,43)(H2,25,28,29)(H2,37,38,39)/t13-,17-,18-,19+,23-/m1/s1 |
|---|
| InChI Key | QAQREVBBADEHPA-IEXPHMLFSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as acyl coas. These are organic compounds containing a coenzyme A substructure linked to an acyl chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | Acyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Aminopyrimidine
- Monoalkyl phosphate
- Alkyl phosphate
- Pyrimidine
- Phosphoric acid ester
- Imidolactam
- Monosaccharide
- N-substituted imidazole
- Organic phosphoric acid derivative
- Tetrahydrofuran
- Azole
- Imidazole
- Heteroaromatic compound
- Thiocarboxylic acid ester
- Secondary alcohol
- Amino acid or derivatives
- Carbothioic s-ester
- Thiocarboxylic acid or derivatives
- Sulfenyl compound
- Propargyl-type 1,3-dipolar organic compound
- Carboximidic acid
- Carboximidic acid derivative
- Carboxylic acid derivative
- Organic 1,3-dipolar compound
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Organopnictogen compound
- Organic oxygen compound
- Organic oxide
- Amine
- Hydrocarbon derivative
- Primary amine
- Carbonyl group
- Alcohol
- Organosulfur compound
- Organic nitrogen compound
- Organonitrogen compound
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatized |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - Propionyl-CoA GC-MS (TMS_1_1) - 70eV, Positive | Not Available | 2021-10-19 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Propionyl-CoA GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-10-19 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Propionyl-CoA GC-MS (TMS_1_3) - 70eV, Positive | Not Available | 2021-10-19 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Propionyl-CoA GC-MS (TMS_1_4) - 70eV, Positive | Not Available | 2021-10-19 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Propionyl-CoA GC-MS (TMS_1_5) - 70eV, Positive | Not Available | 2021-10-19 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Propionyl-CoA GC-MS (TMS_1_6) - 70eV, Positive | Not Available | 2021-10-19 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Propionyl-CoA GC-MS (TMS_1_7) - 70eV, Positive | Not Available | 2021-10-19 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Propionyl-CoA GC-MS (TMS_1_8) - 70eV, Positive | Not Available | 2021-10-19 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionyl-CoA 10V, Positive-QTOF | splash10-000i-1901000220-8d8b91979e9c40176540 | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionyl-CoA 20V, Positive-QTOF | splash10-000i-0912000000-7c91e2c2f2f0d7b4a3ad | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionyl-CoA 40V, Positive-QTOF | splash10-000i-2901000000-4311d5cc2b23055e1e3e | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionyl-CoA 10V, Negative-QTOF | splash10-0a7i-9830230550-4bec7e3f4052a0f39eef | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionyl-CoA 20V, Negative-QTOF | splash10-0560-4910100000-d7281a44fcd4120a6bcd | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionyl-CoA 40V, Negative-QTOF | splash10-057i-6900100000-19dd2ec65951d55b39f7 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionyl-CoA 10V, Negative-QTOF | splash10-00di-0000000190-23b4b60665f8f21035f9 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionyl-CoA 20V, Negative-QTOF | splash10-053r-9000000000-6406a938fedabc06792a | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionyl-CoA 40V, Negative-QTOF | splash10-00kr-7003503900-84f2f16540c6fdc2e156 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionyl-CoA 10V, Positive-QTOF | splash10-00di-0000000090-c1703348da5a9b18dd37 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionyl-CoA 20V, Positive-QTOF | splash10-000i-1911003540-d00d30c93f6f200af707 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionyl-CoA 40V, Positive-QTOF | splash10-014i-0129000000-299d7589b9a24abdcb95 | 2021-09-23 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum |
|
|---|