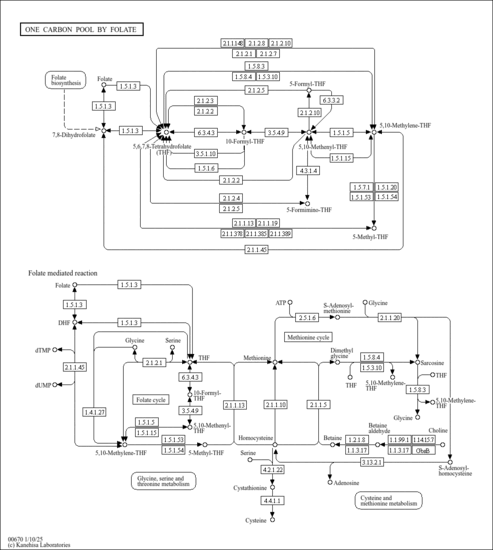
| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| 5,10-Methenyltetrahydrofolic acid,1TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N)=NC4=O)C=C1)C(=O)[O-] | 4728.4 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,1TMS,isomer #2 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)[NH]1 | 4868.0 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,1TMS,isomer #3 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1[NH]C(N)=NC2=O | 4686.8 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,1TMS,isomer #4 | C[Si](C)(C)N1C(N)=NC(=O)C2=C1NC[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]21 | 4771.9 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,1TMS,isomer #5 | C[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 4713.6 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #1 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])C=C4)C=[N+]23)[NH]1 | 4765.1 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #1 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])C=C4)C=[N+]23)[NH]1 | 4643.5 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #1 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])C=C4)C=[N+]23)[NH]1 | 6597.2 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #10 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1N([Si](C)(C)C)C(N)=NC2=O | 4650.5 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #10 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1N([Si](C)(C)C)C(N)=NC2=O | 4595.3 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #10 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1N([Si](C)(C)C)C(N)=NC2=O | 6401.8 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #11 | C[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 4666.9 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #11 | C[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 4429.2 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #11 | C[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 6608.7 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N)=NC4=O)C=C1)[Si](C)(C)C | 4637.9 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N)=NC4=O)C=C1)[Si](C)(C)C | 4474.4 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N)=NC4=O)C=C1)[Si](C)(C)C | 6485.3 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)C(=O)[O-] | 4704.1 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)C(=O)[O-] | 4522.9 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)C(=O)[O-] | 6641.7 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N)=NC4=O)N([Si](C)(C)C)C[C@@H]3C2)C=C1)C(=O)[O-] | 4571.3 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N)=NC4=O)N([Si](C)(C)C)C[C@@H]3C2)C=C1)C(=O)[O-] | 4629.8 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N)=NC4=O)N([Si](C)(C)C)C[C@@H]3C2)C=C1)C(=O)[O-] | 6336.0 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #5 | C[Si](C)(C)N(C1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)[NH]1)[Si](C)(C)C | 4837.5 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #5 | C[Si](C)(C)N(C1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)[NH]1)[Si](C)(C)C | 4638.9 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #5 | C[Si](C)(C)N(C1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)[NH]1)[Si](C)(C)C | 6576.1 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #6 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C | 4803.4 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #6 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C | 4659.2 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #6 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C | 6611.0 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]21 | 4737.4 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]21 | 4708.2 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]21 | 6305.1 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #8 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)[NH]1 | 4754.9 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #8 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)[NH]1 | 4539.2 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #8 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)[NH]1 | 6549.2 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #9 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]2C2=C1[NH]C(N)=NC2=O | 4544.5 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #9 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]2C2=C1[NH]C(N)=NC2=O | 4494.9 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TMS,isomer #9 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]2C2=C1[NH]C(N)=NC2=O | 6285.3 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)C(=O)[O-] | 4715.9 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)C(=O)[O-] | 4619.0 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)C(=O)[O-] | 6271.4 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #10 | C[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 4713.3 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #10 | C[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 4541.5 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #10 | C[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 6227.3 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #11 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C | 4688.3 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #11 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C | 4725.9 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #11 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C | 5955.2 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #12 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C | 4663.4 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #12 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C | 4552.6 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #12 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C | 6271.0 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #13 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]21 | 4581.2 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #13 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]21 | 4596.8 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #13 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]21 | 5963.7 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #14 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]2C2=C1N([Si](C)(C)C)C(N)=NC2=O | 4530.4 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #14 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]2C2=C1N([Si](C)(C)C)C(N)=NC2=O | 4512.4 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #14 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]2C2=C1N([Si](C)(C)C)C(N)=NC2=O | 6116.6 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #2 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C | 4689.6 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #2 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C | 4608.1 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #2 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C | 6312.6 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #3 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])C=C3)C=[N+]21 | 4609.2 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #3 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])C=C3)C=[N+]21 | 4679.3 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #3 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])C=C3)C=[N+]21 | 6012.5 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #4 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)[NH]1 | 4635.1 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #4 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)[NH]1 | 4545.2 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #4 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)[NH]1 | 6226.9 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #5 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)[Si](C)(C)C | 4560.9 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #5 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)[Si](C)(C)C | 4441.3 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #5 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)[Si](C)(C)C | 6339.3 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N)=NC4=O)N([Si](C)(C)C)C[C@@H]3C2)C=C1)[Si](C)(C)C | 4436.0 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N)=NC4=O)N([Si](C)(C)C)C[C@@H]3C2)C=C1)[Si](C)(C)C | 4527.5 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N)=NC4=O)N([Si](C)(C)C)C[C@@H]3C2)C=C1)[Si](C)(C)C | 6019.9 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #7 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C)C[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)C(=O)[O-] | 4567.3 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #7 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C)C[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)C(=O)[O-] | 4601.6 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #7 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C)C[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)C(=O)[O-] | 6168.7 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #8 | C[Si](C)(C)N(C1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C)[Si](C)(C)C | 4786.0 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #8 | C[Si](C)(C)N(C1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C)[Si](C)(C)C | 4639.2 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #8 | C[Si](C)(C)N(C1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C)[Si](C)(C)C | 6281.2 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #9 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4717.7 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #9 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4647.5 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TMS,isomer #9 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 6007.1 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[Si](C)(C)C | 4599.8 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[Si](C)(C)C | 4561.4 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[Si](C)(C)C | 5884.2 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #10 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]2C2=C1[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4578.4 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #10 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]2C2=C1[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4584.3 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #10 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]2C2=C1[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 5663.1 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #11 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C | 4541.6 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #11 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C | 4635.7 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #11 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C | 5624.4 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)C(=O)[O-] | 4667.7 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)C(=O)[O-] | 4597.5 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)C(=O)[O-] | 5960.1 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)N([Si](C)(C)C)C[C@@H]3C2)C=C1)C(=O)[O-] | 4607.1 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)N([Si](C)(C)C)C[C@@H]3C2)C=C1)C(=O)[O-] | 4645.4 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)N([Si](C)(C)C)C[C@@H]3C2)C=C1)C(=O)[O-] | 5712.7 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #4 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C | 4575.1 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #4 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C | 4680.6 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #4 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C | 5662.3 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #5 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C | 4553.2 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #5 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C | 4521.8 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #5 | C[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C | 5951.0 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #6 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]21 | 4463.7 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #6 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]21 | 4579.3 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #6 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]21 | 5662.9 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #7 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C)C[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)[Si](C)(C)C | 4437.5 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #7 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C)C[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)[Si](C)(C)C | 4527.3 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #7 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C)C[C@@H]3C2)N([Si](C)(C)C)C(N)=NC4=O)C=C1)[Si](C)(C)C | 5892.5 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #8 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4691.6 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #8 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4711.9 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #8 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 5640.7 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #9 | C[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 4645.1 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #9 | C[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 4572.9 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,4TMS,isomer #9 | C[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 5917.7 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[Si](C)(C)C | 4577.7 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[Si](C)(C)C | 4544.1 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[Si](C)(C)C | 5625.2 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)N([Si](C)(C)C)C[C@@H]3C2)C=C1)[Si](C)(C)C | 4485.1 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)N([Si](C)(C)C)C[C@@H]3C2)C=C1)[Si](C)(C)C | 4606.2 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)N([Si](C)(C)C)C[C@@H]3C2)C=C1)[Si](C)(C)C | 5392.3 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C)C[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)C(=O)[O-] | 4596.1 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C)C[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)C(=O)[O-] | 4684.1 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C)C[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)C(=O)[O-] | 5369.5 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #4 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C | 4449.8 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #4 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C | 4605.5 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #4 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)[O-])[Si](C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C | 5355.5 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #5 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]2C2=C1N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4566.9 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #5 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]2C2=C1N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4667.4 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,5TMS,isomer #5 | C[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C)C=C3)C=[N+]2C2=C1N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 5346.6 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,6TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C)C[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[Si](C)(C)C | 4497.4 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,6TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C)C[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[Si](C)(C)C | 4656.2 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,6TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C)C[C@@H]3C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC4=O)C=C1)[Si](C)(C)C | 5076.0 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,1TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N)=NC4=O)C=C1)C(=O)[O-] | 5031.0 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,1TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)[NH]1 | 5085.7 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,1TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1[NH]C(N)=NC2=O | 4977.2 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,1TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)N1C(N)=NC(=O)C2=C1NC[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]21 | 5005.9 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,1TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 4980.6 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)[O-])C=C4)C=[N+]23)[NH]1 | 5204.6 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)[O-])C=C4)C=[N+]23)[NH]1 | 5058.7 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)[O-])C=C4)C=[N+]23)[NH]1 | 6456.8 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 5154.5 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 5023.5 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 6325.4 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C(C)(C)C)C(N)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 5153.7 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C(C)(C)C)C(N)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 4878.0 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C(C)(C)C)C(N)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 6445.0 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N)=NC4=O)C=C1)[Si](C)(C)C(C)(C)C | 5153.6 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N)=NC4=O)C=C1)[Si](C)(C)C(C)(C)C | 4899.8 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N)=NC4=O)C=C1)[Si](C)(C)C(C)(C)C | 6376.4 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C(C)(C)C)C(N)=NC4=O)C=C1)C(=O)[O-] | 5178.6 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C(C)(C)C)C(N)=NC4=O)C=C1)C(=O)[O-] | 4953.6 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C(C)(C)C)C(N)=NC4=O)C=C1)C(=O)[O-] | 6480.2 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N)=NC4=O)N([Si](C)(C)C(C)(C)C)C[C@@H]3C2)C=C1)C(=O)[O-] | 5120.2 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N)=NC4=O)N([Si](C)(C)C(C)(C)C)C[C@@H]3C2)C=C1)C(=O)[O-] | 5008.5 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N)=NC4=O)N([Si](C)(C)C(C)(C)C)C[C@@H]3C2)C=C1)C(=O)[O-] | 6288.7 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)N(C1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)[NH]1)[Si](C)(C)C(C)(C)C | 5262.9 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)N(C1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)[NH]1)[Si](C)(C)C(C)(C)C | 5055.1 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)N(C1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)[NH]1)[Si](C)(C)C(C)(C)C | 6437.3 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C | 5233.3 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C | 5078.1 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C | 6388.3 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]21 | 5170.3 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]21 | 5101.4 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]21 | 6254.7 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C4)C=[N+]23)[NH]1 | 5175.6 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C4)C=[N+]23)[NH]1 | 4973.0 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C4)C=[N+]23)[NH]1 | 6411.8 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C3)C=[N+]2C2=C1[NH]C(N)=NC2=O | 5088.5 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C3)C=[N+]2C2=C1[NH]C(N)=NC2=O | 4912.5 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,2TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C3)C=[N+]2C2=C1[NH]C(N)=NC2=O | 6246.5 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC4=O)C=C1)C(=O)[O-] | 5311.9 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC4=O)C=C1)C(=O)[O-] | 5234.1 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC4=O)C=C1)C(=O)[O-] | 6093.4 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 5304.3 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 5178.7 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC4=O)C=C1)[C@@H](CCC(=O)O)C(=O)[O-] | 6056.2 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C | 5285.1 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C | 5306.7 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)C[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C | 5867.8 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #12 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C | 5256.0 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #12 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C | 5167.2 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #12 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C | 6049.6 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #13 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C3)C=[N+]21 | 5184.6 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #13 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C3)C=[N+]21 | 5189.8 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #13 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C3)C=[N+]21 | 5911.5 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #14 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C3)C=[N+]2C2=C1N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 5219.7 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #14 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C3)C=[N+]2C2=C1N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 5131.8 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #14 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C3)C=[N+]2C2=C1N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 6052.3 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C | 5278.1 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C | 5202.9 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C | 6078.0 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)[O-])C=C3)C=[N+]21 | 5204.7 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)[O-])C=C3)C=[N+]21 | 5242.5 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)C[C@@H]1CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)[O-])C=C3)C=[N+]21 | 5938.0 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C4)C=[N+]23)[NH]1 | 5215.1 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C4)C=[N+]23)[NH]1 | 5160.8 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)[O-])[Si](C)(C)C(C)(C)C)C=C4)C=[N+]23)[NH]1 | 6083.3 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C(C)(C)C)C(N)=NC4=O)C=C1)[Si](C)(C)C(C)(C)C | 5236.5 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C(C)(C)C)C(N)=NC4=O)C=C1)[Si](C)(C)C(C)(C)C | 5068.3 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C(NC[C@@H]3C2)N([Si](C)(C)C(C)(C)C)C(N)=NC4=O)C=C1)[Si](C)(C)C(C)(C)C | 6186.3 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N)=NC4=O)N([Si](C)(C)C(C)(C)C)C[C@@H]3C2)C=C1)[Si](C)(C)C(C)(C)C | 5175.4 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N)=NC4=O)N([Si](C)(C)C(C)(C)C)C[C@@H]3C2)C=C1)[Si](C)(C)C(C)(C)C | 5099.6 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)[O-])N(C(=O)C1=CC=C(N2C=[N+]3C4=C([NH]C(N)=NC4=O)N([Si](C)(C)C(C)(C)C)C[C@@H]3C2)C=C1)[Si](C)(C)C(C)(C)C | 5990.4 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C(C)(C)C)C[C@@H]3C2)N([Si](C)(C)C(C)(C)C)C(N)=NC4=O)C=C1)C(=O)[O-] | 5248.3 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C(C)(C)C)C[C@@H]3C2)N([Si](C)(C)C(C)(C)C)C(N)=NC4=O)C=C1)C(=O)[O-] | 5187.1 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N2C=[N+]3C4=C(N([Si](C)(C)C(C)(C)C)C[C@@H]3C2)N([Si](C)(C)C(C)(C)C)C(N)=NC4=O)C=C1)C(=O)[O-] | 6090.0 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)N(C1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 5340.8 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)N(C1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 5251.3 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)N(C1=NC(=O)C2=C(NC[C@@H]3CN(C4=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C4)C=[N+]23)N1[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 6044.6 | Standard polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 5311.8 | Semi standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 5235.2 | Standard non polar | 33892256 |
| 5,10-Methenyltetrahydrofolic acid,3TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)N1C[C@@H]2CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)[O-])C=C3)C=[N+]2C2=C1[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 5915.5 | Standard polar | 33892256 |