| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2006-02-28 12:15:37 UTC |
|---|
| Update Date | 2023-02-21 17:15:55 UTC |
|---|
| HMDB ID | HMDB0001889 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Theophylline |
|---|
| Description | Theophylline, also known as quibron TSR or uniphyl, belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. Theophylline also binds to the adenosine A2B receptor and blocks adenosine mediated bronchoconstriction. Theophylline is a drug which is used for the treatment of the symptoms and reversible airflow obstruction associated with chronic asthma and other chronic lung diseases, such as emphysema and chronic bronchitis. Theophylline is marketed under several brand names such as Theophylline and Theochron, and it is indicated mainly for asthma, bronchospasm, and COPD. Within humans, theophylline participates in a number of enzymatic reactions. In particular, theophylline and formaldehyde can be biosynthesized from caffeine; which is mediated by the enzymes cytochrome P450 1A2, cytochrome P450 3A4, cytochrome P450 2C8, cytochrome P450 2C9, and cytochrome P450 2E1. In addition, theophylline can be converted into 1-methylxanthine and formaldehyde; which is mediated by the enzyme cytochrome P450 1A2. In humans, theophylline is involved in caffeine metabolism. Theophylline is a bitter tasting compound. Outside of the human body, Theophylline is found, on average, in the highest concentration within cocoa beans and tea. Theophylline has also been detected, but not quantified in a few different foods, such as arabica coffee, lemons, and pummelo. This could make theophylline a potential biomarker for the consumption of these foods. Theophylline is a potentially toxic compound. |
|---|
| Structure | CN1C2=C(NC=N2)C(=O)N(C)C1=O InChI=1S/C7H8N4O2/c1-10-5-4(8-3-9-5)6(12)11(2)7(10)13/h3H,1-2H3,(H,8,9) |
|---|
| Synonyms | | Value | Source |
|---|
| 1,3-Dimethyl-7H-purine-2,6-dione | ChEBI | | 1,3-Dimethylxanthine | ChEBI | | Elixophyllin | ChEBI | | Respbid | ChEBI | | Theo-dur | ChEBI | | Theolair | ChEBI | | Theophyllin | ChEBI | | Theophylline anhydrous | ChEBI | | Uniphyl | ChEBI | | Quibron-t | Kegg | | Theo-24 | Kegg | | Theodur g | Kegg | | Accurbron | HMDB | | Acet-theocin | HMDB | | Aerolate | HMDB | | Aerolate III | HMDB | | Aerolate SR | HMDB | | Aminophylline | HMDB | | Aquaphyllin | HMDB | | Armophylline | HMDB | | Asbron | HMDB | | Asmax | HMDB | | Austyn | HMDB | | Bronkodyl | HMDB | | Bronkodyl SR | HMDB | | Choledyl sa | HMDB | | Constant-T | HMDB | | Diphyllin | HMDB | | Doraphyllin | HMDB | | Duraphyl | HMDB | | Dyspne-inhal | HMDB | | Elixex | HMDB | | Elixicon | HMDB | | Elixomin | HMDB | | Elixophyllin SR | HMDB | | Elixophylline | HMDB | | Euphylline | HMDB | | Euphylong | HMDB | | Labid | HMDB | | Lanophyllin | HMDB | | Liquophylline | HMDB | | Liquorice | HMDB | | Maphylline | HMDB | | Medaphyllin | HMDB | | Nuelin | HMDB | | Optiphyllin | HMDB | | Parkophyllin | HMDB | | Pseudotheophylline | HMDB | | Quibron t/sr | HMDB | | Quibron-t/sr | HMDB | | Slo-bid | HMDB | | Slo-phyllin | HMDB | | Solosin | HMDB | | Somophyllin-CRT | HMDB | | Somophyllin-DF | HMDB | | Somophyllin-T | HMDB | | Spophyllin retard | HMDB | | Sustaire | HMDB | | Synophylate | HMDB | | Synophylate-l.a. cenules | HMDB | | T-Phyl | HMDB | | Tefamin | HMDB | | Teofilina | HMDB | | Teofyllamin | HMDB | | Teolair | HMDB | | Theacitin | HMDB | | Theal tabl. | HMDB | | Theal tablets | HMDB | | Theo-dur-sprinkle | HMDB | | Theobid | HMDB | | Theobid jr. | HMDB | | Theochron | HMDB | | Theocin | HMDB | | Theoclair-SR | HMDB | | Theoclear 80 | HMDB | | Theoclear l.a.-130 | HMDB | | Theoclear la | HMDB | | Theoclear-200 | HMDB | | Theoclear-80 | HMDB | | Theocontin | HMDB | | Theodel | HMDB | | Theofol | HMDB | | Theograd | HMDB | | Theolair-SR | HMDB | | Theolix | HMDB | | Theolixir | HMDB | | Theona p | HMDB | | Theophyl | HMDB | | Theophyl-225 | HMDB | | Theophyl-SR | HMDB | | Theophyline | HMDB | | Theophylline-SR | HMDB | | Theostat 80 | HMDB | | Theovent | HMDB | | Uni-dur | HMDB | | Unifyl | HMDB | | Uniphyllin | HMDB | | Xanthium | HMDB | | Xantivent | HMDB | | Aerobin | HMDB | | Bronchoparat | HMDB | | Glycine theophyllinate | HMDB | | Nuelin s.a. | HMDB | | Slo phyllin | HMDB | | SloPhyllin | HMDB | | Somophyllin T | HMDB | | Theo24 | HMDB | | Theoconfin continuous | HMDB | | Theonite | HMDB | | CT Arzneimittel brand OF theophylline sodium glycinate | HMDB | | Von CT, theo | HMDB | | Mundipharma brand OF theophylline sodium glycinate | HMDB | | Quibron T SR | HMDB | | Quibron T-SR | HMDB | | Sodium glycinate, theophylline | HMDB | | Theo 24 | HMDB | | Theophyllinate, glycine | HMDB | | Theophylline sodium glycinate | HMDB | | Theospan | HMDB | | Uniphylline | HMDB | | CT-Arzneimittel brand OF theophylline sodium glycinate | HMDB | | ConstantT | HMDB | | Fameasan brand OF theophylline sodium glycinate | HMDB | | Lodrane | HMDB | | Monospan | HMDB | | Quibron TSR | HMDB | | Theodur | HMDB | | Theon | HMDB | | Theopek | HMDB | | Theostat | HMDB | | CT, Theo von | HMDB | | Theo von CT | HMDB | | 1,3 Dimethylxanthine | HMDB | | 3,7-Dihydro-1,3-dimethyl-1H-purine-2,6-dione | HMDB | | Afonilum retard | HMDB | | Anhydrous, theophylline | HMDB | | Constant T | HMDB | | Fujisawa brand OF theophylline sodium glycinate | HMDB | | Glycinate, theophylline sodium | HMDB | | SomophyllinT | HMDB | | Theo dur | HMDB |
|
|---|
| Chemical Formula | C7H8N4O2 |
|---|
| Average Molecular Weight | 180.164 |
|---|
| Monoisotopic Molecular Weight | 180.06472552 |
|---|
| IUPAC Name | 1,3-dimethyl-2,3,6,7-tetrahydro-1H-purine-2,6-dione |
|---|
| Traditional Name | theophylline |
|---|
| CAS Registry Number | 58-55-9 |
|---|
| SMILES | CN1C2=C(NC=N2)C(=O)N(C)C1=O |
|---|
| InChI Identifier | InChI=1S/C7H8N4O2/c1-10-5-4(8-3-9-5)6(12)11(2)7(10)13/h3H,1-2H3,(H,8,9) |
|---|
| InChI Key | ZFXYFBGIUFBOJW-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Xanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthine
- 6-oxopurine
- Purinone
- Alkaloid or derivatives
- Pyrimidone
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Vinylogous amide
- Lactam
- Urea
- Azacycle
- Hydrocarbon derivative
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 273 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 7.36 mg/mL at 25 °C | Not Available | | LogP | -0.02 | HANSCH,C ET AL. (1995) |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Theophylline,1TMS,isomer #1 | CN1C(=O)C2=C(N=CN2[Si](C)(C)C)N(C)C1=O | 1945.6 | Semi standard non polar | 33892256 | | Theophylline,1TMS,isomer #1 | CN1C(=O)C2=C(N=CN2[Si](C)(C)C)N(C)C1=O | 2069.2 | Standard non polar | 33892256 | | Theophylline,1TMS,isomer #1 | CN1C(=O)C2=C(N=CN2[Si](C)(C)C)N(C)C1=O | 2632.4 | Standard polar | 33892256 | | Theophylline,1TBDMS,isomer #1 | CN1C(=O)C2=C(N=CN2[Si](C)(C)C(C)(C)C)N(C)C1=O | 2181.8 | Semi standard non polar | 33892256 | | Theophylline,1TBDMS,isomer #1 | CN1C(=O)C2=C(N=CN2[Si](C)(C)C(C)(C)C)N(C)C1=O | 2251.7 | Standard non polar | 33892256 | | Theophylline,1TBDMS,isomer #1 | CN1C(=O)C2=C(N=CN2[Si](C)(C)C(C)(C)C)N(C)C1=O | 2667.4 | Standard polar | 33892256 |
|
|---|
| Spectra |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - Theophylline GC-MS (1 TMS) | splash10-0f79-6970000000-224461ad62a44dbdf860 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Theophylline EI-B (Non-derivatized) | splash10-001j-7900000000-a08735d528e738752429 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Theophylline CI-B (Non-derivatized) | splash10-001i-0900000000-d0882f7d959c726e7623 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Theophylline EI-B (Non-derivatized) | splash10-001i-9700000000-8d0e1898a6571fbca10a | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Theophylline GC-MS (Non-derivatized) | splash10-0f79-6970000000-224461ad62a44dbdf860 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Theophylline GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fka-4900000000-878cab6882fd80efba3b | 2017-07-27 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Theophylline GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-00lr-9500000000-2c8464c2fe84464c207f | 2014-09-20 | Not Available | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline Quattro_QQQ 10V, Positive-QTOF (Annotated) | splash10-00di-0900000000-0092516012d6a2a93b31 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline Quattro_QQQ 25V, Positive-QTOF (Annotated) | splash10-00di-4900000000-dcf52c18a6996f412b1a | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline Quattro_QQQ 40V, Positive-QTOF (Annotated) | splash10-00kf-9000000000-09089337909892ef44cb | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline EI-B (Unknown) , Positive-QTOF | splash10-001j-7900000000-a08735d528e738752429 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline CI-B (Unknown) , Positive-QTOF | splash10-001i-0900000000-d0882f7d959c726e7623 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative-QTOF | splash10-004i-0900000000-c4943571126a44bb9e5a | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative-QTOF | splash10-004i-0900000000-bc12ce29acd02fa749b8 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative-QTOF | splash10-03k9-0900000000-d63f60043f186fbb9bc0 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative-QTOF | splash10-070l-4900000000-d293ab2fa6199dbaf97a | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative-QTOF | splash10-05ru-9400000000-dde4775588d52c83806f | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative-QTOF | splash10-004i-0900000000-556e382f583d610ef1f0 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative-QTOF | splash10-004i-0900000000-89e34f5158856ba33469 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative-QTOF | splash10-03k9-0900000000-65a6897d72954e875b00 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative-QTOF | splash10-070l-4900000000-df16604bd6c40cee8f24 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative-QTOF | splash10-05r3-9500000000-39a721dcecc86ed95507 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive-QTOF | splash10-001i-1900000000-e67ff7ef9a955b90eb3f | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive-QTOF | splash10-001i-3900000000-1870952d98dbba22ace8 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive-QTOF | splash10-00di-7900000000-dc0e9606776c43a7823f | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive-QTOF | splash10-014j-9300000000-2c4c6490dda3ed3b563f | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive-QTOF | splash10-014i-9000000000-26a6c0c23a465afdcde1 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive-QTOF | splash10-01q9-1900000000-b11ca3441bb29fef1906 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive-QTOF | splash10-0002-9400000000-63726fe0b49a9dc6ba7e | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive-QTOF | splash10-00di-9100000000-139f765bc9b5855960c6 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive-QTOF | splash10-00di-9000000000-88b5cb393b960973ac64 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Theophylline LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive-QTOF | splash10-00yi-9000000000-1ebbddce3c7133972546 | 2012-08-31 | HMDB team, MONA | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
|
|---|
| Biological Properties |
|---|
| Cellular Locations | - Cytoplasm (predicted from logP)
|
|---|
| Biospecimen Locations | - Blood
- Cerebrospinal Fluid (CSF)
- Feces
- Urine
|
|---|
| Tissue Locations | - Adipose Tissue
- Adrenal Medulla
- Brain
- Epidermis
- Fibroblasts
- Kidney
- Leukocyte
- Liver
- Neuron
- Pancreas
- Placenta
- Platelet
- Skeletal Muscle
- Testis
|
|---|
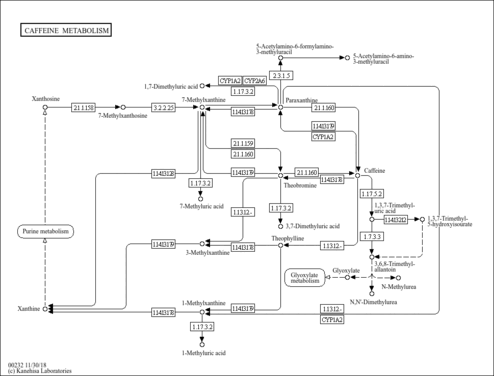
| Pathways | |
|---|
| Normal Concentrations |
|---|
| |
| Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 29.0 +/- 11.0 uM | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 0.18 (0.00-0.73) umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details |
|
|---|
| Abnormal Concentrations |
|---|
| |
| Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal adenoma | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 0.166 +/- 0.0522 uM | Adult (>18 years old) | Not Specified | Favorable outcome from traumatic brain injury | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 0.0911 +/- 0.0205 uM | Adult (>18 years old) | Not Specified | Traumatic brain injury | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 0.105 +/- 0.0183 uM | Adult (>18 years old) | Not Specified | Traumatic brain injury (TBI) | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal adenoma | | details | | Urine | Detected and Quantified | 7.0 (3.2-11.0) umol/mmol creatinine | Adult (>18 years old) | Both | Asthma | | details |
|
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | | Traumatic brain injury |
|---|
- Sachse KT, Jackson EK, Wisniewski SR, Gillespie DG, Puccio AM, Clark RS, Dixon CE, Kochanek PM: Increases in cerebrospinal fluid caffeine concentration are associated with favorable outcome after severe traumatic brain injury in humans. J Cereb Blood Flow Metab. 2008 Feb;28(2):395-401. Epub 2007 Aug 8. [PubMed:17684518 ]
| | Head injury |
|---|
- Sachse KT, Jackson EK, Wisniewski SR, Gillespie DG, Puccio AM, Clark RS, Dixon CE, Kochanek PM: Increases in cerebrospinal fluid caffeine concentration are associated with favorable outcome after severe traumatic brain injury in humans. J Cereb Blood Flow Metab. 2008 Feb;28(2):395-401. Epub 2007 Aug 8. [PubMed:17684518 ]
| | Colorectal cancer |
|---|
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
| | Asthma |
|---|
- Zydron M, Baranowski J, Baranowska I: Separation, pre-concentration, and HPLC analysis of methylxanthines in urine samples. J Sep Sci. 2004 Oct;27(14):1166-72. [PubMed:15537072 ]
|
|
|---|
| Associated OMIM IDs | |
|---|
| External Links |
|---|
| DrugBank ID | DB00277 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB000453 |
|---|
| KNApSAcK ID | C00001510 |
|---|
| Chemspider ID | 2068 |
|---|
| KEGG Compound ID | C07130 |
|---|
| BioCyc ID | CPD-12479 |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Theophylline |
|---|
| METLIN ID | 1458 |
|---|
| PubChem Compound | 2153 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 28177 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | Not Available |
|---|
| MarkerDB ID | MDB00000353 |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| General References | - Bailey DN: Relative binding of acetaminophen, lidocaine, phenobarbital, phenytoin, quinidine, and theophylline to human tissues in vitro. J Anal Toxicol. 1997 Jan-Feb;21(1):1-4. [PubMed:9013284 ]
- Bath PM: Theophylline, aminophylline, caffeine and analogues for acute ischaemic stroke. Cochrane Database Syst Rev. 2004;(3):CD000211. [PubMed:15266427 ]
- Yasui K, Agematsu K, Shinozaki K, Hokibara S, Nagumo H, Yamada S, Kobayashi N, Komiyama A: Effects of theophylline on human eosinophil functions: comparative study with neutrophil functions. J Leukoc Biol. 2000 Aug;68(2):194-200. [PubMed:10947063 ]
- Gerbershagen MU, Fiege M, Weisshorn R, Kolodzie K, Wappler F: [Theophylline induces contractures in porcine skeletal muscle preparations with the disposition to malignant hyperthermia]. Anasthesiol Intensivmed Notfallmed Schmerzther. 2004 Mar;39(3):147-52. [PubMed:15042504 ]
- Mahomed AG, Theron AJ, Anderson R, Feldman C: Anti-oxidative effects of theophylline on human neutrophils involve cyclic nucleotides and protein kinase A. Inflammation. 1998 Dec;22(6):545-57. [PubMed:9824770 ]
- Andreas S, Reiter H, Luthje L, Delekat A, Grunewald RW, Hasenfuss G, Somers VK: Differential effects of theophylline on sympathetic excitation, hemodynamics, and breathing in congestive heart failure. Circulation. 2004 Oct 12;110(15):2157-62. Epub 2004 Oct 4. [PubMed:15466632 ]
- Yano Y, Yoshida M, Hoshino S, Inoue K, Kida H, Yanagita M, Takimoto T, Hirata H, Kijima T, Kumagai T, Osaki T, Tachibana I, Kawase I: Anti-fibrotic effects of theophylline on lung fibroblasts. Biochem Biophys Res Commun. 2006 Mar 17;341(3):684-90. Epub 2006 Jan 18. [PubMed:16430859 ]
- Mohiuddin AA, Bath FJ, Bath PM: Theophylline, aminophylline, caffeine and analogues for acute ischaemic stroke. Cochrane Database Syst Rev. 2000;(2):CD000211. [PubMed:10796327 ]
- Mehta R, Weinberger B, Usmani SS, Wapnir RA, Harper RG: Theophylline alters neutrophil function in preterm infants. Biol Neonate. 2002;81(3):176-81. [PubMed:11937723 ]
- Benoehr P, Krueth P, Bokemeyer C, Grenz A, Osswald H, Hartmann JT: Nephroprotection by theophylline in patients with cisplatin chemotherapy: a randomized, single-blinded, placebo-controlled trial. J Am Soc Nephrol. 2005 Feb;16(2):452-8. Epub 2004 Dec 8. [PubMed:15590762 ]
- Spoelstra FM, Berends C, Dijkhuizen B, de Monchy JG, Kauffman HF: Effect of theophylline on CD11b and L-selectin expression and density of eosinophils and neutrophils in vitro. Eur Respir J. 1998 Sep;12(3):585-91. [PubMed:9762784 ]
- Teplinskaia LE, Filichkina NS, Matevosova KS: [Efficiency of treatment of uveitis with the drug superlymph]. Vestn Oftalmol. 2005 Jul-Aug;121(4):22-6. [PubMed:16223038 ]
- Yoshiike T, Aikawa Y, Sindhvananda J, Suto H, Nishimura K, Kawamoto T, Ogawa H: Skin barrier defect in atopic dermatitis: increased permeability of the stratum corneum using dimethyl sulfoxide and theophylline. J Dermatol Sci. 1993 Apr;5(2):92-6. [PubMed:8357787 ]
- Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. [PubMed:32033212 ]
|
|---|