| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2006-05-22 14:17:37 UTC |
|---|
| Update Date | 2023-02-21 17:16:11 UTC |
|---|
| HMDB ID | HMDB0002123 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 1,3,7-Trimethyluric acid |
|---|
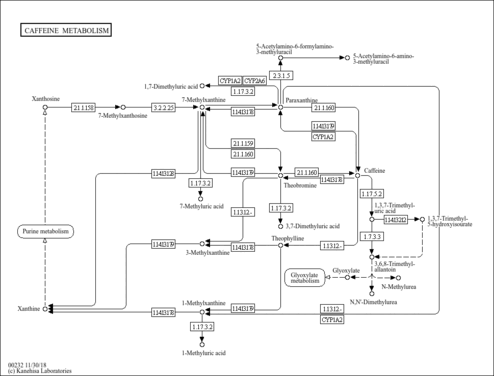
| Description | 1,3,7-Trimethyluric acid is a methyl derivative of uric acid, found occasionally in human urine. 1,3,7-Trimethyluracil is one of the purine components in urinary calculi. Methylated purines originate from the metabolism of methylxanthines (caffeine, theophylline and theobromine). Methyluric acids are indistinguishable from uric acid by simple methods routinely used in clinical laboratories, requiring the use of high-performance liquid chromatography (HPLC). Purine derivatives in urinary calculi could be considered markers of abnormal purine metabolism. The content of a purine derivative in stone depends on its average urinary excretion in the general population, similarity to the chemical structure of uric acid, and content of the latter in stone. This suggests that purines in stones represent a solid solution with uric acid as solvent. It is also plausible that methylxanthines, ubiquitous components of the diet and drugs, are involved in the pathogenesis of urolithiasis. Caffeine is metabolized via successive pathways mainly catalyzed by CYP1A2, xanthine oxidase or N-acetyltransferase-2 to give 14 different metabolites. CYP1A2 activity shows an inter-individual variability among the population. CYP1A2, an isoform of the CYP1A cytochrome P450 super-family, is involved in the metabolism of many drugs and plays a potentially important role in the induction of chemical carcinogenesis. (PMID:11712316 , 15833286 , 3506820 , 15013152 ). |
|---|
| Structure | CN1C(=O)NC2=C1C(=O)N(C)C(=O)N2C InChI=1S/C8H10N4O3/c1-10-4-5(9-7(10)14)11(2)8(15)12(3)6(4)13/h1-3H3,(H,9,14) |
|---|
| Synonyms | | Value | Source |
|---|
| 1,3,7-Trimethyl-2,3,6,7,8,9-hexahydro-1H-purine-2,6,8-trione | ChEBI | | 1,3,7-Trimethylurate | ChEBI | | 7,9-Dihydro-1,3,7-trimethyl-1H-purine-2,6,8(3H)-trione | ChEBI | | 8-Oxocaffeine | ChEBI | | 8-Oxy-caffeine | ChEBI | | 1,3,7-Trimethylate | Generator | | 1,3,7-Trimethylic acid | Generator | | 1,3, 7-Trimethyluric acid | HMDB | | 1,3,7-Trimethyl-7,9-dihydro-1H-purine-2,6,8(3H)-trione | HMDB | | 2,6,8-Trihydroxy-1,3,7-trimethylpurine | HMDB | | 8-Hydroxy-1,3,7-trimethyl-3,7-dihydro-1H-purine-2,6-dione | HMDB | | Trimethyl uric acid | HMDB |
|
|---|
| Chemical Formula | C8H10N4O3 |
|---|
| Average Molecular Weight | 210.19 |
|---|
| Monoisotopic Molecular Weight | 210.075290206 |
|---|
| IUPAC Name | 1,3,7-trimethyl-2,3,6,7,8,9-hexahydro-1H-purine-2,6,8-trione |
|---|
| Traditional Name | 1,3,7-trimethyluric acid |
|---|
| CAS Registry Number | 5415-44-1 |
|---|
| SMILES | CN1C(=O)NC2=C1C(=O)N(C)C(=O)N2C |
|---|
| InChI Identifier | InChI=1S/C8H10N4O3/c1-10-4-5(9-7(10)14)11(2)8(15)12(3)6(4)13/h1-3H3,(H,9,14) |
|---|
| InChI Key | BYXCFUMGEBZDDI-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Xanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthine
- 6-oxopurine
- Purinone
- Alkaloid or derivatives
- Pyrimidone
- N-substituted imidazole
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Vinylogous amide
- Lactam
- Urea
- Azacycle
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 5.5 mg/mL at 15 °C | Not Available | | LogP | -0.37 | GASPARI,F & BONATI,M (1987) |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| 1,3,7-Trimethyluric acid,1TMS,isomer #1 | CN1C(=O)C2=C(N(C)C1=O)N([Si](C)(C)C)C(=O)N2C | 1967.6 | Semi standard non polar | 33892256 | | 1,3,7-Trimethyluric acid,1TMS,isomer #1 | CN1C(=O)C2=C(N(C)C1=O)N([Si](C)(C)C)C(=O)N2C | 2235.1 | Standard non polar | 33892256 | | 1,3,7-Trimethyluric acid,1TMS,isomer #1 | CN1C(=O)C2=C(N(C)C1=O)N([Si](C)(C)C)C(=O)N2C | 2791.4 | Standard polar | 33892256 | | 1,3,7-Trimethyluric acid,1TBDMS,isomer #1 | CN1C(=O)C2=C(N(C)C1=O)N([Si](C)(C)C(C)(C)C)C(=O)N2C | 2180.0 | Semi standard non polar | 33892256 | | 1,3,7-Trimethyluric acid,1TBDMS,isomer #1 | CN1C(=O)C2=C(N(C)C1=O)N([Si](C)(C)C(C)(C)C)C(=O)N2C | 2441.1 | Standard non polar | 33892256 | | 1,3,7-Trimethyluric acid,1TBDMS,isomer #1 | CN1C(=O)C2=C(N(C)C1=O)N([Si](C)(C)C(C)(C)C)C(=O)N2C | 2761.1 | Standard polar | 33892256 |
|
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - 1,3,7-Trimethyluric acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-0udi-1900000000-7ceff209b1b0d4f92cf0 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 1,3,7-Trimethyluric acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid Quattro_QQQ 10V, N/A-QTOF (Annotated) | splash10-03di-0090000000-c6489ab09ec74614db4c | 2012-07-25 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid Quattro_QQQ 25V, N/A-QTOF (Annotated) | splash10-001a-6900000000-ac2e74a460c865c0636b | 2012-07-25 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid Quattro_QQQ 40V, N/A-QTOF (Annotated) | splash10-067l-9100000000-cbac42dd288a8ef44fd8 | 2012-07-25 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid Linear Ion Trap , negative-QTOF | splash10-0006-0900000000-a9bff7d94cd5ffe280b9 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid Linear Ion Trap , positive-QTOF | splash10-0udj-0900000000-5316637dfdcce19f087a | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 20V, Positive-QTOF | splash10-0imj-3930000000-0593f0aaec496efc7696 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 30V, Positive-QTOF | splash10-000f-9500000000-fb28fd8903d55811b0a1 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 20V, Negative-QTOF | splash10-000f-0910000000-eca560d36d14fb72dfe9 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 10V, Negative-QTOF | splash10-0a4i-0190000000-6d4b9f5e8e42f3040e5e | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 40V, Negative-QTOF | splash10-000l-7900000000-2ba0d4f876fceab5182f | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 40V, Negative-QTOF | splash10-000f-9700000000-a42f3584a2bad7add22d | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 20V, Negative-QTOF | splash10-052o-1920000000-53b778bbd3181f2ed83a | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 30V, Negative-QTOF | splash10-000l-3900000000-4305cd286c077e521079 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 10V, Negative-QTOF | splash10-0a4i-0690000000-62dc7eb029289b61c3b1 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 35V, Negative-QTOF | splash10-000f-0900000000-3eb9fdcfd2e67d0f4e2c | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 40V, Positive-QTOF | splash10-0006-9100000000-f438ad2b70b2e9735643 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 10V, Positive-QTOF | splash10-03di-0190000000-42add8be62bbafed200c | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 20V, Positive-QTOF | splash10-03dj-2930000000-2855ca36942d5e8f069e | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 40V, Positive-QTOF | splash10-066u-9200000000-708fd95bbe618c2ace7d | 2021-09-20 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 10V, Positive-QTOF | splash10-03di-0090000000-153ea9ea8b4fe3692bfb | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 20V, Positive-QTOF | splash10-0ik9-1790000000-2d17afbf34659471f524 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 40V, Positive-QTOF | splash10-0m2a-4900000000-c783d40d7ed0c6ae44a9 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 10V, Negative-QTOF | splash10-0a4i-0190000000-9858aab04a40f6286f93 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 20V, Negative-QTOF | splash10-0a4i-1790000000-2089332ab35983ee2c77 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1,3,7-Trimethyluric acid 40V, Negative-QTOF | splash10-066u-8900000000-53f14da0e8f43679f03f | 2015-05-27 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | 2012-12-05 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
|
|---|
| Disease References | | Colorectal cancer |
|---|
- Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ: Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016 Mar 25;11(3):e0152126. doi: 10.1371/journal.pone.0152126. eCollection 2016. [PubMed:27015276 ]
- Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP: Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016 Jun 6;4:11. doi: 10.1186/s40170-016-0151-y. eCollection 2016. [PubMed:27275383 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
| | Asthma |
|---|
- Zydron M, Baranowski J, Baranowska I: Separation, pre-concentration, and HPLC analysis of methylxanthines in urine samples. J Sep Sci. 2004 Oct;27(14):1166-72. [PubMed:15537072 ]
|
|
|---|