| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2006-05-22 14:17:39 UTC |
|---|
| Update Date | 2022-03-07 02:49:13 UTC |
|---|
| HMDB ID | HMDB0002177 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Cis-8,11,14,17-Eicosatetraenoic acid |
|---|
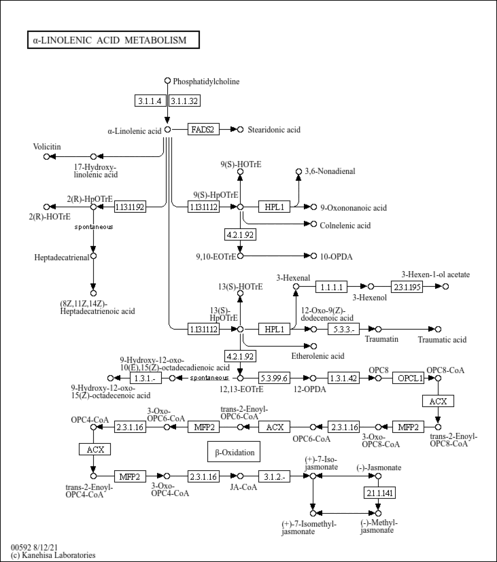
| Description | Cis-8,11,14,17-Eicosatetraenoic acid is an eicosanoid present in marine lipids, a minor n-3 polyunsaturated fatty acid (PUFA) which is a position isomer of 20:4n-6. n-3 PUFA contained in marine lipids appear to have a protective effect against coronary heart disease and thrombosis. Human platelets metabolize 8,11,14,17-eicosatetraenoic acid primarily into 12-hydroxy-8,10,14,17-eicosatetraenoic acid. The eicosanoids are a diverse family of molecules that have powerful effects on cell function. They are best known as intercellular messengers, having autocrine and paracrine effects following their secretion from the cells that synthesize them. The diversity of possible products that can be synthesized from eicosatrienoic acid is due, in part to the variety of enzymes that can act on it. Studies have placed many, but not all, of these enzymes at or inside the nucleus. In some cases, the nuclear import or export of eicosatrienoic acid-processing enzymes is highly regulated. Furthermore, nuclear receptors that are activated by specific eicosanoids are known to exist. Taken together, these findings indicate that the enzymatic conversion of eicosatrienoic acid to specific signaling molecules can occur in the nucleus, that it is regulated, and that the synthesized products may act within the nucleus. PMID: 3109494 , 8142566 , 16574479 , 15896193 , 10037447 ). Trans fatty acids are characteristically produced during industrial hydrogenation of plant oils. |
|---|
| Structure | CC\C=C/C\C=C/C\C=C/C\C=C/CCCCCCC(O)=O InChI=1S/C20H32O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18-19-20(21)22/h3-4,6-7,9-10,12-13H,2,5,8,11,14-19H2,1H3,(H,21,22)/b4-3-,7-6-,10-9-,13-12- |
|---|
| Synonyms | | Value | Source |
|---|
| (8Z,11Z,14Z,17Z)-Eicosa-8,11,14,17-tetraenoic acid | ChEBI | | (8Z,11Z,14Z,17Z)-Eicosatetraenoic acid | ChEBI | | (8Z,11Z,14Z,17Z)-Icosatetraenoic acid | ChEBI | | (Z,Z,Z,Z)-Eicosa-8,11,14,17-tetraenoic acid | ChEBI | | (Z,Z,Z,Z)-Icosa-8,11,14,17-tetraenoic acid | ChEBI | | 8Z,11Z,14Z,17Z-Eicosatetraenoic acid | ChEBI | | all-cis-8,11,14,17-Eicosatetraenoic acid | ChEBI | | all-Z-8,11,14,17-Eicosatetraenoic acid | ChEBI | | all-Z-8,11,14,17-Icosatetraenoic acid | ChEBI | | ETA | ChEBI | | Omega-3-arachidonic acid | ChEBI | | (8Z,11Z,14Z,17Z)-Eicosa-8,11,14,17-tetraenoate | Generator | | (8Z,11Z,14Z,17Z)-Eicosatetraenoate | Generator | | (8Z,11Z,14Z,17Z)-Icosatetraenoate | Generator | | (Z,Z,Z,Z)-Eicosa-8,11,14,17-tetraenoate | Generator | | (Z,Z,Z,Z)-Icosa-8,11,14,17-tetraenoate | Generator | | 8Z,11Z,14Z,17Z-Eicosatetraenoate | Generator | | all-cis-8,11,14,17-Eicosatetraenoate | Generator | | all-Z-8,11,14,17-Eicosatetraenoate | Generator | | all-Z-8,11,14,17-Icosatetraenoate | Generator | | Omega-3-arachidonate | Generator | | cis-8,11,14,17-Eicosatetraenoate | Generator | | 8, 11, 14, 17-Icosatetraenoate | HMDB | | 8, 11, 14, 17-Icosatetraenoic acid | HMDB | | 8,11,14,17-Eicosatetraenoate | HMDB | | 8,11,14,17-Eicosatetraenoic acid | HMDB | | 8,11,14,17-Eicosatetraenoic acid, Z-isomer | HMDB |
|
|---|
| Chemical Formula | C20H32O2 |
|---|
| Average Molecular Weight | 304.4669 |
|---|
| Monoisotopic Molecular Weight | 304.240230268 |
|---|
| IUPAC Name | (8Z,11Z,14Z,17Z)-icosa-8,11,14,17-tetraenoic acid |
|---|
| Traditional Name | (8Z,11Z,14Z,17Z)-icosa-8,11,14,17-tetraenoic acid |
|---|
| CAS Registry Number | 2091-26-1 |
|---|
| SMILES | CC\C=C/C\C=C/C\C=C/C\C=C/CCCCCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C20H32O2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18-19-20(21)22/h3-4,6-7,9-10,12-13H,2,5,8,11,14-19H2,1H3,(H,21,22)/b4-3-,7-6-,10-9-,13-12- |
|---|
| InChI Key | HQPCSDADVLFHHO-LTKCOYKYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as long-chain fatty acids. These are fatty acids with an aliphatic tail that contains between 13 and 21 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Long-chain fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Long-chain fatty acid
- Unsaturated fatty acid
- Straight chain fatty acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | Not Available |
|---|
| Physical Properties |
|---|
| State | Liquid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Cis-8,11,14,17-Eicosatetraenoic acid,1TMS,isomer #1 | CC/C=C\C/C=C\C/C=C\C/C=C\CCCCCCC(=O)O[Si](C)(C)C | 2385.2 | Semi standard non polar | 33892256 | | Cis-8,11,14,17-Eicosatetraenoic acid,1TBDMS,isomer #1 | CC/C=C\C/C=C\C/C=C\C/C=C\CCCCCCC(=O)O[Si](C)(C)C(C)(C)C | 2627.8 | Semi standard non polar | 33892256 |
|
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-000l-6790000000-9f82428f04530524e4d2 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid GC-MS (1 TMS) - 70eV, Positive | splash10-01w0-7962000000-84eacc6d7a7ceee0c99a | 2017-10-06 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid Linear Ion Trap , negative-QTOF | splash10-0a4i-0090000000-ac340d2fdec196225551 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid Linear Ion Trap , positive-QTOF | splash10-002f-0591000000-9c83feea251cbef18bac | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid Linear Ion Trap , positive-QTOF | splash10-059m-0491000000-c78942db68649dad3705 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid 10V, Positive-QTOF | splash10-052r-0092000000-adcd16a168a2136833fa | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid 20V, Positive-QTOF | splash10-0a4r-4691000000-e3736249247ab6a17228 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid 40V, Positive-QTOF | splash10-052n-8950000000-af9e1166c7a23305f645 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid 10V, Negative-QTOF | splash10-0udi-0039000000-057a58eb03c1d61d43f9 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid 20V, Negative-QTOF | splash10-0zg0-1095000000-a5e878ae7122108346ea | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid 40V, Negative-QTOF | splash10-0a4l-9240000000-d349b1db215c158d4b05 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid 10V, Negative-QTOF | splash10-0udi-0019000000-e301ae96559ee65daa0e | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid 20V, Negative-QTOF | splash10-0udr-2069000000-b0db6033219193d0d9e4 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid 40V, Negative-QTOF | splash10-006x-9320000000-58917ba5f6a987214949 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid 10V, Positive-QTOF | splash10-0a4r-4796000000-26f15a5b60f20cad946e | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid 20V, Positive-QTOF | splash10-001l-7910000000-dcb9693160ae6a462cd8 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Cis-8,11,14,17-Eicosatetraenoic acid 40V, Positive-QTOF | splash10-0036-9300000000-a07065b94af0ab13e426 | 2021-09-24 | Wishart Lab | View Spectrum |
|
|---|