| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2012-09-11 21:07:53 UTC |
|---|
| Update Date | 2022-03-07 02:54:46 UTC |
|---|
| HMDB ID | HMDB0036062 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Ubiquinone 6 |
|---|
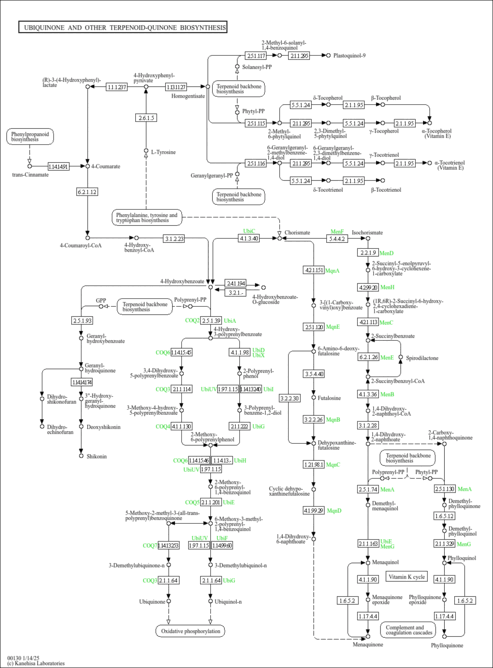
| Description | Ubiquinone-6 is a member of the chemical class known as Polyprenylbenzoquinones. These are compounds containing a polyisoprene chain attached to a quinone at the second ring position. Ubiquione-6 has just 6 isoprene units. Normally in humans it has 10. Ubiquinone-6 is an intermediate in the synthesis of Ubiquionone 10. It is an endogenouse compound but it has also been isolated from foods containing baker's yeast. Ubiquionone 10 (CoQ10) is involved in cellular respiration. It is fat-soluble and is therefore mobile in cellular membranes; it plays a unique role in the electron transport chain (ETC). In the inner bacterial membrane, electrons from NADH and succinate pass through the ETC to the oxygen, which is then reduced to water. The transfer of electrons through ETC results in the pumping of H+ across the membrane creating a proton gradient across the membrane, which is used by ATP synthase (located on the membrane) to generate ATP. |
|---|
| Structure | COC1=C(OC)C(=O)C(CC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)C)=C(C)C1=O InChI=1S/C39H58O4/c1-28(2)16-11-17-29(3)18-12-19-30(4)20-13-21-31(5)22-14-23-32(6)24-15-25-33(7)26-27-35-34(8)36(40)38(42-9)39(43-10)37(35)41/h16,18,20,22,24,26H,11-15,17,19,21,23,25,27H2,1-10H3 |
|---|
| Synonyms | | Value | Source |
|---|
| Coenzyme Q6 | HMDB | | Coenzyme QQ6 | HMDB | | Coenzyme-Q6 | HMDB | | CoQ-6 | HMDB | | CoQ6 | HMDB | | Ubiquinone 30 | HMDB | | Ubiquinone Q6 | HMDB | | Ubiquinone(6) | HMDB | | UBIQUINONE-6 | HMDB |
|
|---|
| Chemical Formula | C39H58O4 |
|---|
| Average Molecular Weight | 590.8754 |
|---|
| Monoisotopic Molecular Weight | 590.433510344 |
|---|
| IUPAC Name | 2-(3,7,11,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl)-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione |
|---|
| Traditional Name | 2-(3,7,11,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl)-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione |
|---|
| CAS Registry Number | 1065-31-2 |
|---|
| SMILES | COC1=C(OC)C(=O)C(CC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)CCC=C(C)C)=C(C)C1=O |
|---|
| InChI Identifier | InChI=1S/C39H58O4/c1-28(2)16-11-17-29(3)18-12-19-30(4)20-13-21-31(5)22-14-23-32(6)24-15-25-33(7)26-27-35-34(8)36(40)38(42-9)39(43-10)37(35)41/h16,18,20,22,24,26H,11-15,17,19,21,23,25,27H2,1-10H3 |
|---|
| InChI Key | GXNFPEOUKFOTKY-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as ubiquinones. These are coenzyme Q derivatives containing a 5, 6-dimethoxy-3-methyl(1,4-benzoquinone) moiety to which an isoprenyl group is attached at ring position 2(or 6). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Quinone and hydroquinone lipids |
|---|
| Direct Parent | Ubiquinones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Polyprenylbenzoquinone
- Sesterterpenoid
- Ubiquinone skeleton
- Quinone
- P-benzoquinone
- Vinylogous ester
- Cyclic ketone
- Ketone
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Liquid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 19 - 20 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatized |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - Ubiquinone 6 GC-MS (Non-derivatized) - 70eV, Positive | splash10-0fi9-2268490000-d4a46fb7bbfe567b3bc9 | 2017-09-01 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 10V, Positive-QTOF | splash10-0007-0112190000-0e3f9b8afdf1ec72101d | 2015-04-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 20V, Positive-QTOF | splash10-002b-0649330000-25374eb6b7e2528209f9 | 2015-04-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 40V, Positive-QTOF | splash10-00kb-2125910000-2353dc21b36ad0226438 | 2015-04-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 10V, Negative-QTOF | splash10-000i-0000090000-888a7e1c1887cd6289a7 | 2015-04-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 20V, Negative-QTOF | splash10-000i-0000090000-cdb5f8e03d488aabf72f | 2015-04-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 40V, Negative-QTOF | splash10-0079-9100580000-0ddcd7334398c6806be9 | 2015-04-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 10V, Positive-QTOF | splash10-0007-0212190000-d6790c276a238e59aeca | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 20V, Positive-QTOF | splash10-002b-0649330000-25374eb6b7e2528209f9 | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 40V, Positive-QTOF | splash10-00mk-2125910000-fd96f37265811c6c3531 | 2015-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 10V, Negative-QTOF | splash10-000i-0000090000-1e48e9465d0a2b5e046c | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 20V, Negative-QTOF | splash10-000i-0100090000-308b58d9d2d89e2a0150 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 40V, Negative-QTOF | splash10-0079-8100590000-fa8ea854e31b895398f6 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 10V, Positive-QTOF | splash10-000e-4619380000-15b4ea76379fc8709a54 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 20V, Positive-QTOF | splash10-0002-4915010000-9f9f91ee19707f8366d2 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 40V, Positive-QTOF | splash10-000y-8924000000-472232f8797340057410 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 10V, Negative-QTOF | splash10-000i-0000090000-652b47e044f4b53fb940 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 20V, Negative-QTOF | splash10-07br-0611090000-96454b02ed13c80d5269 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Ubiquinone 6 40V, Negative-QTOF | splash10-003r-1694370000-5e9b03bff5e8c4cb6b63 | 2021-09-22 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-23 | Wishart Lab | View Spectrum |
|
|---|