| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2023-05-30 20:56:03 UTC |
|---|
| HMDB ID | HMDB0000237 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Propionic acid |
|---|
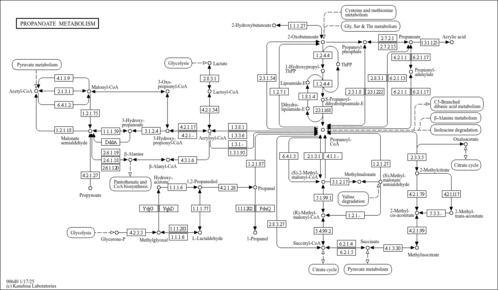
| Description | Propionic acid (PA) is an organic acid. It exists a clear liquid with a pungent and unpleasant smell somewhat resembling body odor. Propionic acid (PA) is widely used as an antifungal agent in food. It is present naturally at low levels in dairy products and occurs ubiquitously, together with other short-chain fatty acids (SCFA), in the gastro-intestinal tract of humans and other mammals as an end-product of the microbial digestion of carbohydrates. The metabolism of propionic acid begins with its conversion to propionyl coenzyme A, the usual first step in the metabolism of carboxylic acids. Since propionic acid has three carbons, propionyl-CoA cannot directly enter either beta oxidation or the citric acid cycles. In most vertebrates, propionyl-CoA is carboxylated to D-methylmalonyl-CoA, which is isomerised to L-methylmalonyl-CoA. Propionic acid has significant physiological activity in animals. Propionic acid is irritant but produces no acute systemic effects and has no demonstrable genotoxic potential (PMID 1628870 ). The human skin is host of several species of bacteria known as Propionibacteria, which are named after their ability to produce propionic acid. The most notable one is the Cutibacterium acnes (formerly known as Propionibacterium acnes), which lives mainly in the sebaceous glands of the skin and is one of the principal causes of acne. Propionic aciduria is one of the most frequent organic acidurias, a disease that comprise many various disorders. The outcome of patients born with Propionic aciduria is poor intellectual development patterns, with 60% having an IQ less than 75 and requiring special education. Successful liver and/or renal transplantations, in a few patients, have resulted in better quality of life but have not necessarily prevented neurological and various visceral complications. These results emphasize the need for permanent metabolic follow-up whatever the therapeutic strategy (PMID 15868474 ). Decreased early mortality, less severe symptoms at diagnosis, and more favorable short-term neurodevelopmental outcome were recorded in patients identified through expanded newborn screening. (PMID 16763906 )↵ When propionic acid is infused directly into rodents' brains, it produces hyperactivity, dystonia, social impairment, perseveration and brain changes (e.g., innate neuroinflammation, glutathione depletion) that may be used as a means to model autism in rats. Propionic acid is a metabolite of Bacteroides, Clostridium, Dialister, Megasphaera, Phascolarctobacterium, Propionibacterium, Propionigenum, Salmonella, Selenomonas and Veillonella (https://www.mdpi.com/2311-5637/3/2/21). |
|---|
| Structure | InChI=1S/C3H6O2/c1-2-3(4)5/h2H2,1H3,(H,4,5) |
|---|
| Synonyms | | Value | Source |
|---|
| Acide propanoique | ChEBI | | Acide propionique | ChEBI | | Carboxyethane | ChEBI | | CH3-CH2-COOH | ChEBI | | Ethanecarboxylic acid | ChEBI | | Ethylformic acid | ChEBI | | Metacetonic acid | ChEBI | | Methylacetic acid | ChEBI | | PA | ChEBI | | Propanoic acid | ChEBI | | Propioic acid | ChEBI | | Propionsaeure | ChEBI | | Propoic acid | ChEBI | | Pseudoacetic acid | ChEBI | | Propionate | Kegg | | Ethanecarboxylate | Generator | | Ethylformate | Generator | | Metacetonate | Generator | | Methylacetate | Generator | | Propanoate | Generator | | Propioate | Generator | | Propoate | Generator | | Pseudoacetate | Generator | | Adofeed | HMDB | | Antischim b | HMDB | | Luprosil | HMDB | | MonoProp | HMDB | | Propanate | HMDB | | Propcorn | HMDB | | Propkorn | HMDB | | Prozoin | HMDB | | Toxi-check | HMDB | | Propionic acid, zinc salt | HMDB | | Potassium propionate | HMDB | | Chromium propionate | HMDB | | Lithium propanoate | HMDB | | Zinc propionate | HMDB |
|
|---|
| Chemical Formula | C3H6O2 |
|---|
| Average Molecular Weight | 74.0785 |
|---|
| Monoisotopic Molecular Weight | 74.036779436 |
|---|
| IUPAC Name | propanoic acid |
|---|
| Traditional Name | propanoic acid |
|---|
| CAS Registry Number | 79-09-4 |
|---|
| SMILES | CCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C3H6O2/c1-2-3(4)5/h2H2,1H3,(H,4,5) |
|---|
| InChI Key | XBDQKXXYIPTUBI-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as carboxylic acids. Carboxylic acids are compounds containing a carboxylic acid group with the formula -C(=O)OH. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Carboxylic acids |
|---|
| Direct Parent | Carboxylic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Monocarboxylic acid or derivatives
- Carboxylic acid
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Liquid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | -20.7 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 1000 mg/mL | Not Available | | LogP | 0.33 | HANSCH,C ET AL. (1995) |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - Propionic acid EI-B (Non-derivatized) | splash10-004i-9000000000-51f674be972a6c17185b | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Propionic acid EI-B (Non-derivatized) | splash10-004i-9000000000-691dcd080b30c9898350 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Propionic acid EI-B (Non-derivatized) | splash10-004i-9000000000-51f674be972a6c17185b | 2018-05-18 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Propionic acid EI-B (Non-derivatized) | splash10-004i-9000000000-691dcd080b30c9898350 | 2018-05-18 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Propionic acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-00b9-9000000000-abf322c73e6badb078bf | 2016-09-22 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Propionic acid GC-MS (1 TMS) - 70eV, Positive | splash10-00fr-9100000000-cbfe9e32208652e70047 | 2017-10-06 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Propionic acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Propionic acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Propionic acid GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-00b9-9000000000-0bb3297c4159bed2316e | 2014-09-20 | Not Available | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid Quattro_QQQ 10V, Positive-QTOF (Annotated) | splash10-004i-9000000000-1af60fc458a7f351a9b0 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid Quattro_QQQ 25V, Positive-QTOF (Annotated) | splash10-004i-9000000000-aa6e765fc867ac8be641 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid Quattro_QQQ 40V, Positive-QTOF (Annotated) | splash10-0006-9000000000-27e0b790e192d1304449 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid EI-B (HITACHI RMU-6M) , Positive-QTOF | splash10-004i-9000000000-51f674be972a6c17185b | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid EI-B (HITACHI M-80B) , Positive-QTOF | splash10-004i-9000000000-90d9e0181596093a2f85 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative-QTOF | splash10-00di-9000000000-bdd7baa3d1bda886fb77 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative-QTOF | splash10-00di-9000000000-e73379c8765802cf3228 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative-QTOF | splash10-00di-9000000000-d6832c04c8b2ca0fdfa3 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative-QTOF | splash10-00di-9000000000-d0c93844dbfaed791bb0 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid LC-ESI-QQ , negative-QTOF | splash10-00di-9000000000-bdd7baa3d1bda886fb77 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid LC-ESI-QQ , negative-QTOF | splash10-00di-9000000000-e73379c8765802cf3228 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid LC-ESI-QQ , negative-QTOF | splash10-00di-9000000000-d6832c04c8b2ca0fdfa3 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Propionic acid LC-ESI-QQ , negative-QTOF | splash10-00di-9000000000-d0c93844dbfaed791bb0 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionic acid 10V, Positive-QTOF | splash10-0a4i-9000000000-47364fadf00a5a2b7e93 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionic acid 20V, Positive-QTOF | splash10-0a6r-9000000000-76fad523c005a6510264 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionic acid 40V, Positive-QTOF | splash10-0a6r-9000000000-a1c0234da57ff32c6e12 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionic acid 10V, Positive-QTOF | splash10-0a4i-9000000000-47364fadf00a5a2b7e93 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionic acid 20V, Positive-QTOF | splash10-0a6r-9000000000-76fad523c005a6510264 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionic acid 40V, Positive-QTOF | splash10-0a6r-9000000000-a1c0234da57ff32c6e12 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionic acid 10V, Negative-QTOF | splash10-00di-9000000000-db59da781a70634d2526 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionic acid 20V, Negative-QTOF | splash10-05fr-9000000000-bea4ff21e6ab6c664412 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionic acid 40V, Negative-QTOF | splash10-0a4i-9000000000-782832f8f5ab85f2ef4f | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionic acid 10V, Negative-QTOF | splash10-00di-9000000000-db59da781a70634d2526 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionic acid 20V, Negative-QTOF | splash10-05fr-9000000000-bea4ff21e6ab6c664412 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Propionic acid 40V, Negative-QTOF | splash10-0a4i-9000000000-782832f8f5ab85f2ef4f | 2015-05-27 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Experimental 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | 2021-10-10 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
|
|---|
| Disease References | | Propionic acidemia |
|---|
- Thompson GN, Chalmers RA, Walter JH, Bresson JL, Lyonnet SL, Reed PJ, Saudubray JM, Leonard JV, Halliday D: The use of metronidazole in management of methylmalonic and propionic acidaemias. Eur J Pediatr. 1990 Aug;149(11):792-6. [PubMed:2226555 ]
| | Methylmalonic acidemia |
|---|
- Thompson GN, Chalmers RA, Walter JH, Bresson JL, Lyonnet SL, Reed PJ, Saudubray JM, Leonard JV, Halliday D: The use of metronidazole in management of methylmalonic and propionic acidaemias. Eur J Pediatr. 1990 Aug;149(11):792-6. [PubMed:2226555 ]
| | Colorectal cancer |
|---|
- Monleon D, Morales JM, Barrasa A, Lopez JA, Vazquez C, Celda B: Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed. 2009 Apr;22(3):342-8. doi: 10.1002/nbm.1345. [PubMed:19006102 ]
- Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP: Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013 Aug 6;8(8):e70803. doi: 10.1371/journal.pone.0070803. Print 2013. [PubMed:23940645 ]
- Lin Y, Ma C, Liu C, Wang Z, Yang J, Liu X, Shen Z, Wu R: NMR-based fecal metabolomics fingerprinting as predictors of earlier diagnosis in patients with colorectal cancer. Oncotarget. 2016 May 17;7(20):29454-64. doi: 10.18632/oncotarget.8762. [PubMed:27107423 ]
- Wang X, Wang J, Rao B, Deng L: Gut flora profiling and fecal metabolite composition of colorectal cancer patients and healthy individuals. Exp Ther Med. 2017 Jun;13(6):2848-2854. doi: 10.3892/etm.2017.4367. Epub 2017 Apr 20. [PubMed:28587349 ]
| | Irritable bowel syndrome |
|---|
- Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A: Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res. 2011 Sep 2;10(9):4208-18. doi: 10.1021/pr2003598. Epub 2011 Aug 8. [PubMed:21761941 ]
- Walton C, Fowler DP, Turner C, Jia W, Whitehead RN, Griffiths L, Dawson C, Waring RH, Ramsden DB, Cole JA, Cauchi M, Bessant C, Hunter JO: Analysis of volatile organic compounds of bacterial origin in chronic gastrointestinal diseases. Inflamm Bowel Dis. 2013 Sep;19(10):2069-78. doi: 10.1097/MIB.0b013e31829a91f6. [PubMed:23867873 ]
| | Ulcerative colitis |
|---|
- Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A: Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res. 2011 Sep 2;10(9):4208-18. doi: 10.1021/pr2003598. Epub 2011 Aug 8. [PubMed:21761941 ]
- Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, Wilson ID, Wang Y: Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007 Feb;6(2):546-51. [PubMed:17269711 ]
- Walton C, Fowler DP, Turner C, Jia W, Whitehead RN, Griffiths L, Dawson C, Waring RH, Ramsden DB, Cole JA, Cauchi M, Bessant C, Hunter JO: Analysis of volatile organic compounds of bacterial origin in chronic gastrointestinal diseases. Inflamm Bowel Dis. 2013 Sep;19(10):2069-78. doi: 10.1097/MIB.0b013e31829a91f6. [PubMed:23867873 ]
- Bjerrum JT, Wang Y, Hao F, Coskun M, Ludwig C, Gunther U, Nielsen OH: Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn's disease and healthy individuals. Metabolomics. 2015;11:122-133. Epub 2014 Jun 1. [PubMed:25598765 ]
- Ahmed I, Greenwood R, Costello B, Ratcliffe N, Probert CS: Investigation of faecal volatile organic metabolites as novel diagnostic biomarkers in inflammatory bowel disease. Aliment Pharmacol Ther. 2016 Mar;43(5):596-611. doi: 10.1111/apt.13522. Epub 2016 Jan 25. [PubMed:26806034 ]
| | Nonalcoholic fatty liver disease |
|---|
- Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, Bailey J, Myers RP, Rioux KP: Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013 Jul;11(7):868-75.e1-3. doi: 10.1016/j.cgh.2013.02.015. Epub 2013 Feb 27. [PubMed:23454028 ]
| | Celiac disease |
|---|
- Di Cagno R, De Angelis M, De Pasquale I, Ndagijimana M, Vernocchi P, Ricciuti P, Gagliardi F, Laghi L, Crecchio C, Guerzoni ME, Gobbetti M, Francavilla R: Duodenal and faecal microbiota of celiac children: molecular, phenotype and metabolome characterization. BMC Microbiol. 2011 Oct 4;11:219. doi: 10.1186/1471-2180-11-219. [PubMed:21970810 ]
| | Crohn's disease |
|---|
- Walton C, Fowler DP, Turner C, Jia W, Whitehead RN, Griffiths L, Dawson C, Waring RH, Ramsden DB, Cole JA, Cauchi M, Bessant C, Hunter JO: Analysis of volatile organic compounds of bacterial origin in chronic gastrointestinal diseases. Inflamm Bowel Dis. 2013 Sep;19(10):2069-78. doi: 10.1097/MIB.0b013e31829a91f6. [PubMed:23867873 ]
- Bjerrum JT, Wang Y, Hao F, Coskun M, Ludwig C, Gunther U, Nielsen OH: Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn's disease and healthy individuals. Metabolomics. 2015;11:122-133. Epub 2014 Jun 1. [PubMed:25598765 ]
- Ahmed I, Greenwood R, Costello B, Ratcliffe N, Probert CS: Investigation of faecal volatile organic metabolites as novel diagnostic biomarkers in inflammatory bowel disease. Aliment Pharmacol Ther. 2016 Mar;43(5):596-611. doi: 10.1111/apt.13522. Epub 2016 Jan 25. [PubMed:26806034 ]
| | Autism |
|---|
- De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, Cristofori F, Guerzoni ME, Gobbetti M, Francavilla R: Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013 Oct 9;8(10):e76993. doi: 10.1371/journal.pone.0076993. eCollection 2013. [PubMed:24130822 ]
| | Pervasive developmental disorder not otherwise specified |
|---|
- De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, Cristofori F, Guerzoni ME, Gobbetti M, Francavilla R: Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013 Oct 9;8(10):e76993. doi: 10.1371/journal.pone.0076993. eCollection 2013. [PubMed:24130822 ]
| | Rheumatoid arthritis |
|---|
- Tie-juan ShaoZhi-xing HeZhi-jun XieHai-chang LiMei-jiao WangCheng-ping Wen. Characterization of ankylosing spondylitis and rheumatoid arthritis using 1H NMR-based metabolomics of human fecal extracts. Metabolomics. April 2016, 12:70 [Link]
| | Eosinophilic esophagitis |
|---|
- Slae, M., Huynh, H., Wishart, D.S. (2014). Analysis of 30 normal pediatric urine samples via NMR spectroscopy (unpublished work). NA.
|
|
|---|
| General References | - Chandler RJ, Aswani V, Tsai MS, Falk M, Wehrli N, Stabler S, Allen R, Sedensky M, Kazazian HH, Venditti CP: Propionyl-CoA and adenosylcobalamin metabolism in Caenorhabditis elegans: evidence for a role of methylmalonyl-CoA epimerase in intermediary metabolism. Mol Genet Metab. 2006 Sep-Oct;89(1-2):64-73. Epub 2006 Jul 14. [PubMed:16843692 ]
- Silwood CJ, Lynch E, Claxson AW, Grootveld MC: 1H and (13)C NMR spectroscopic analysis of human saliva. J Dent Res. 2002 Jun;81(6):422-7. [PubMed:12097436 ]
- Somerma S, Lassus A, Salde L: Assessment of atrophy of human skin caused by corticosteroids using chamber occlusion and suction blister techniques. Acta Derm Venereol. 1984;64(1):41-5. [PubMed:6203280 ]
- Marala RB, Brown JA, Kong JX, Tracey WR, Knight DR, Wester RT, Sun D, Kennedy SP, Hamanaka ES, Ruggeri RB, Hill RJ: Zoniporide: a potent and highly selective inhibitor of human Na(+)/H(+) exchanger-1. Eur J Pharmacol. 2002 Sep 6;451(1):37-41. [PubMed:12223226 ]
- Lin SC, Bergles DE: Synaptic signaling between neurons and glia. Glia. 2004 Aug 15;47(3):290-8. [PubMed:15252819 ]
- Alekseev OM, Widgren EE, Richardson RT, O'Rand MG: Association of NASP with HSP90 in mouse spermatogenic cells: stimulation of ATPase activity and transport of linker histones into nuclei. J Biol Chem. 2005 Jan 28;280(4):2904-11. Epub 2004 Nov 8. [PubMed:15533935 ]
- Robertson MD, Bickerton AS, Dennis AL, Vidal H, Frayn KN: Insulin-sensitizing effects of dietary resistant starch and effects on skeletal muscle and adipose tissue metabolism. Am J Clin Nutr. 2005 Sep;82(3):559-67. [PubMed:16155268 ]
- Esposito BP, Faljoni-Alario A, de Menezes JF, de Brito HF, Najjar R: A circular dichroism and fluorescence quenching study of the interactions between rhodium(II) complexes and human serum albumin. J Inorg Biochem. 1999 May 30;75(1):55-61. [PubMed:10402677 ]
- Jeng JH, Chan CP, Ho YS, Lan WH, Hsieh CC, Chang MC: Effects of butyrate and propionate on the adhesion, growth, cell cycle kinetics, and protein synthesis of cultured human gingival fibroblasts. J Periodontol. 1999 Dec;70(12):1435-42. [PubMed:10632518 ]
- Christensen JK, Varming T, Ahring PK, Jorgensen TD, Nielsen EO: In vitro characterization of 5-carboxyl-2,4-di-benzamidobenzoic acid (NS3763), a noncompetitive antagonist of GLUK5 receptors. J Pharmacol Exp Ther. 2004 Jun;309(3):1003-10. Epub 2004 Feb 25. [PubMed:14985418 ]
- Bintvihok A, Kositcharoenkul S: Effect of dietary calcium propionate on performance, hepatic enzyme activities and aflatoxin residues in broilers fed a diet containing low levels of aflatoxin B1. Toxicon. 2006 Jan;47(1):41-6. Epub 2005 Nov 18. [PubMed:16298407 ]
- Mayer B, Schumacher M, Brandstatter H, Wagner FS, Hermetter A: High-throughput fluorescence screening of antioxidative capacity in human serum. Anal Biochem. 2001 Oct 15;297(2):144-53. [PubMed:11673881 ]
- De Kanter R, De Jager MH, Draaisma AL, Jurva JU, Olinga P, Meijer DK, Groothuis GM: Drug-metabolizing activity of human and rat liver, lung, kidney and intestine slices. Xenobiotica. 2002 May;32(5):349-62. [PubMed:12065058 ]
- Ridge BD, Batt MD, Palmer HE, Jarrett A: The dansyl chloride technique for stratum corneum renewal as an indicator of changes in epidermal mitotic activity following topical treatment. Br J Dermatol. 1988 Feb;118(2):167-74. [PubMed:3348963 ]
- Koeppe RA, Frey KA, Snyder SE, Meyer P, Kilbourn MR, Kuhl DE: Kinetic modeling of N-[11C]methylpiperidin-4-yl propionate: alternatives for analysis of an irreversible positron emission tomography trace for measurement of acetylcholinesterase activity in human brain. J Cereb Blood Flow Metab. 1999 Oct;19(10):1150-63. [PubMed:10532640 ]
- Nguyen TB, Snyder SE, Kilbourn MR: Syntheses of carbon-11 labeled piperidine esters as potential in vivo substrates for acetylcholinesterase. Nucl Med Biol. 1998 Nov;25(8):761-8. [PubMed:9863564 ]
- Wendel U, Zass R, Leupold D: Contribution of odd-numbered fatty acid oxidation to propionate production in neonates with methylmalonic and propionic acidaemias. Eur J Pediatr. 1993 Dec;152(12):1021-3. [PubMed:8131803 ]
- Beutler KT, Pankewycz O, Brautigan DL: Equivalent uptake of organic and inorganic zinc by monkey kidney fibroblasts, human intestinal epithelial cells, or perfused mouse intestine. Biol Trace Elem Res. 1998 Jan;61(1):19-31. [PubMed:9498328 ]
- Harrison PT: Propionic acid and the phenomenon of rodent forestomach tumorigenesis: a review. Food Chem Toxicol. 1992 Apr;30(4):333-40. [PubMed:1628870 ]
- de Baulny HO, Benoist JF, Rigal O, Touati G, Rabier D, Saudubray JM: Methylmalonic and propionic acidaemias: management and outcome. J Inherit Metab Dis. 2005;28(3):415-23. [PubMed:15868474 ]
- Dionisi-Vici C, Deodato F, Roschinger W, Rhead W, Wilcken B: 'Classical' organic acidurias, propionic aciduria, methylmalonic aciduria and isovaleric aciduria: long-term outcome and effects of expanded newborn screening using tandem mass spectrometry. J Inherit Metab Dis. 2006 Apr-Jun;29(2-3):383-9. [PubMed:16763906 ]
|
|---|