| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2006-08-13 00:43:18 UTC |
|---|
| Update Date | 2022-03-07 02:49:19 UTC |
|---|
| HMDB ID | HMDB0003571 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Lauroyl-CoA |
|---|
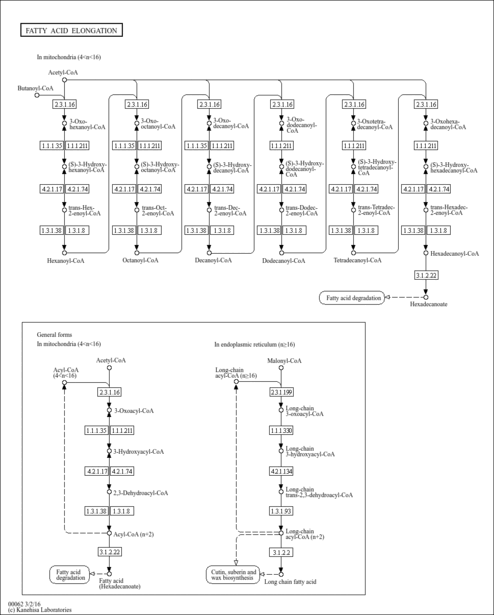
| Description | Lauroyl-CoA, also known as dodecanoyl-CoA or C12:0-CoA, belongs to the class of organic compounds known as beta hydroxy acids and derivatives. Beta hydroxy acids and derivatives are compounds containing a carboxylic acid substituted with a hydroxyl group on the C3 carbon atom. Thus, lauroyl-CoA is considered to be a fatty ester lipid molecule. A medium-chain fatty acyl-CoA that results from the formal condensation of the thiol group of coenzyme A with the carboxy group of lauric (dodecanoic) acid. Lauroyl-CoA is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. Lauroyl-CoA exists in all living species, ranging from bacteria to humans. In humans, lauroyl-CoA is involved in the metabolic disorder called the mitochondrial beta-oxidation of long chain saturated fatty acids pathway. Outside of the human body, Lauroyl-CoA has been detected, but not quantified in, several different foods, such as oval-leaf huckleberries, guava, bamboo shoots, red bell peppers, and calabash. This could make lauroyl-CoA a potential biomarker for the consumption of these foods. |
|---|
| Structure | CCCCCCCCCCCC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N InChI=1S/C33H58N7O17P3S/c1-4-5-6-7-8-9-10-11-12-13-24(42)61-17-16-35-23(41)14-15-36-31(45)28(44)33(2,3)19-54-60(51,52)57-59(49,50)53-18-22-27(56-58(46,47)48)26(43)32(55-22)40-21-39-25-29(34)37-20-38-30(25)40/h20-22,26-28,32,43-44H,4-19H2,1-3H3,(H,35,41)(H,36,45)(H,49,50)(H,51,52)(H2,34,37,38)(H2,46,47,48)/t22-,26-,27-,28+,32-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| C12:0-CoA | ChEBI | | coenzyme A, S-Dodecanoate | ChEBI | | Dodecanoyl-CoA | ChEBI | | Dodecanoyl-coenzyme A | ChEBI | | Lauroyl coenzyme A | ChEBI | | Lauroyl-coenzyme A | ChEBI | | coenzyme A, S-Dodecanoic acid | Generator |
|
|---|
| Chemical Formula | C33H58N7O17P3S |
|---|
| Average Molecular Weight | 949.837 |
|---|
| Monoisotopic Molecular Weight | 949.282273691 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-2-({[({[(3R)-3-[(2-{[2-(dodecanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]-3-hydroxy-2,2-dimethylpropoxy](hydroxy)phosphoryl}oxy)(hydroxy)phosphoryl]oxy}methyl)-4-hydroxyoxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | lauroyl-coa |
|---|
| CAS Registry Number | 6244-92-4 |
|---|
| SMILES | CCCCCCCCCCCC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C33H58N7O17P3S/c1-4-5-6-7-8-9-10-11-12-13-24(42)61-17-16-35-23(41)14-15-36-31(45)28(44)33(2,3)19-54-60(51,52)57-59(49,50)53-18-22-27(56-58(46,47)48)26(43)32(55-22)40-21-39-25-29(34)37-20-38-30(25)40/h20-22,26-28,32,43-44H,4-19H2,1-3H3,(H,35,41)(H,36,45)(H,49,50)(H,51,52)(H2,34,37,38)(H2,46,47,48)/t22-,26-,27-,28+,32-/m1/s1 |
|---|
| InChI Key | YMCXGHLSVALICC-GMHMEAMDSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as beta hydroxy acids and derivatives. Beta hydroxy acids and derivatives are compounds containing a carboxylic acid substituted with a hydroxyl group on the C3 carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Hydroxy acids and derivatives |
|---|
| Sub Class | Beta hydroxy acids and derivatives |
|---|
| Direct Parent | Beta hydroxy acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Beta-hydroxy acid
- Monocyclic benzene moiety
- Morpholine
- Oxazinane
- Piperidine
- N-alkylpyrrolidine
- Benzenoid
- Pyrrolidine
- Amino acid or derivatives
- Carboxylic acid ester
- Tertiary amine
- Tertiary aliphatic amine
- Oxacycle
- Organoheterocyclic compound
- Azacycle
- Carboxylic acid derivative
- Dialkyl ether
- Oxirane
- Ether
- Monocarboxylic acid or derivatives
- Primary alcohol
- Organopnictogen compound
- Organonitrogen compound
- Organooxygen compound
- Amine
- Organic oxide
- Carbonyl group
- Organic oxygen compound
- Organic nitrogen compound
- Alcohol
- Hydrocarbon derivative
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Not Available | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesNot Available |
|---|