| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2023-05-30 20:56:02 UTC |
|---|
| HMDB ID | HMDB0000056 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | beta-Alanine |
|---|
| Description | beta-Alanine is the only naturally occurring beta-amino acid - an amino acid in which the amino group is at the beta-position from the carboxylate group. It is formed in vivo by the degradation of dihydrouracil and carnosine. It is a component of the naturally occurring peptides carnosine and anserine and also of pantothenic acid (vitamin B-5), which itself is a component of coenzyme A. Under normal conditions, beta-alanine is metabolized into acetic acid. beta-Alanine can undergo a transanimation reaction with pyruvate to form malonate-semialdehyde and L-alanine. The malonate semialdehyde can then be converted into malonate via malonate-semialdehyde dehydrogenase. Malonate is then converted into malonyl-CoA and enter fatty acid biosynthesis. Since neuronal uptake and neuronal receptor sensitivity to beta-alanine have been demonstrated, beta-alanine may act as a false transmitter replacing gamma-aminobutyric acid. When present in sufficiently high levels, beta-alanine can act as a neurotoxin, a mitochondrial toxin, and a metabotoxin. A neurotoxin is a compound that damages the brain or nerve tissue. A mitochondrial toxin is a compound that damages mitochondria and reduces cellular respiration as well as oxidative phosphorylation. A metabotoxin is an endogenously produced metabolite that causes adverse health effects at chronically high levels. Chronically high levels of beta-alanine are associated with at least three inborn errors of metabolism, including GABA-transaminase deficiency, hyper-beta-alaninemia, and methylmalonate semialdehyde dehydrogenase deficiency. beta-Alanine is a central nervous system (CNS) depressant and is an inhibitor of GABA transaminase. The associated inhibition of GABA transaminase and displacement of GABA from CNS binding sites can also lead to GABAuria (high levels of GABA in the urine) and convulsions. In addition to its neurotoxicity, beta-alanine reduces cellular levels of taurine, which are required for normal respiratory chain function. Cellular taurine depletion is known to reduce respiratory function and elevate mitochondrial superoxide generation, which damages mitochondria and increases oxidative stress (PMID: 27023909 ). Individuals suffering from mitochondrial defects or mitochondrial toxicity typically develop neurotoxicity, hypotonia, respiratory distress, and cardiac failure. beta-Alanine is a biomarker for the consumption of meat, especially red meat. |
|---|
| Structure | InChI=1S/C3H7NO2/c4-2-1-3(5)6/h1-2,4H2,(H,5,6) |
|---|
| Synonyms | | Value | Source |
|---|
| 3-Aminopropanoic acid | ChEBI | | 3-Aminopropionic acid | ChEBI | | BAla | ChEBI | | beta-Aminopropionic acid | ChEBI | | H-beta-Ala-OH | ChEBI | | Omega-aminopropionic acid | ChEBI | | 3-Aminopropanoate | Kegg | | Abufene | Kegg | | 3-Aminopropionate | Generator | | b-Aminopropionate | Generator | | b-Aminopropionic acid | Generator | | beta-Aminopropionate | Generator | | Β-aminopropionate | Generator | | Β-aminopropionic acid | Generator | | H-b-Ala-OH | Generator | | H-Β-ala-OH | Generator | | Omega-aminopropionate | Generator | | b-Alanine | Generator | | Β-alanine | Generator | | 2-Carboxyethylamine | HMDB | | 3-Amino-propanoate | HMDB | | 3-Amino-propanoic acid | HMDB | | b-Aminopropanoate | HMDB | | b-Aminopropanoic acid | HMDB | | beta Alanine | HMDB | | beta-Aminopropanoate | HMDB | | beta-Aminopropanoic acid | HMDB | | 3 Aminopropionic acid | HMDB | | β-Aminopropanoic acid | PhytoBank | | omega-Aminopropanoic acid | PhytoBank | | ω-Aminopropanoic acid | PhytoBank | | ω-Aminopropionic acid | PhytoBank | | beta-Alanine | PhytoBank |
|
|---|
| Chemical Formula | C3H7NO2 |
|---|
| Average Molecular Weight | 89.0932 |
|---|
| Monoisotopic Molecular Weight | 89.047678473 |
|---|
| IUPAC Name | 3-aminopropanoic acid |
|---|
| Traditional Name | β alanine |
|---|
| CAS Registry Number | 107-95-9 |
|---|
| SMILES | NCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C3H7NO2/c4-2-1-3(5)6/h1-2,4H2,(H,5,6) |
|---|
| InChI Key | UCMIRNVEIXFBKS-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as beta amino acids and derivatives. These are amino acids having a (-NH2) group attached to the beta carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Beta amino acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Beta amino acid or derivatives
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Amine
- Organic oxide
- Hydrocarbon derivative
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Primary aliphatic amine
- Organic oxygen compound
- Carbonyl group
- Organic nitrogen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 200 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 545 mg/mL | Not Available | | LogP | -3.05 | TSAI,RS ET AL. (1991) |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 0.99 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 8.0363 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 7.8 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 352.0 seconds | 40023050 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 427.2 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 319.5 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 62.0 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 223.3 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 81.3 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 258.4 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 228.8 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 814.5 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 530.0 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 34.9 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 595.1 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 209.3 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 291.2 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 725.4 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 506.5 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 360.7 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| beta-Alanine,1TMS,isomer #1 | C[Si](C)(C)OC(=O)CCN | 1022.6 | Semi standard non polar | 33892256 | | beta-Alanine,1TMS,isomer #2 | C[Si](C)(C)NCCC(=O)O | 1172.6 | Semi standard non polar | 33892256 | | beta-Alanine,2TMS,isomer #1 | C[Si](C)(C)NCCC(=O)O[Si](C)(C)C | 1212.1 | Semi standard non polar | 33892256 | | beta-Alanine,2TMS,isomer #1 | C[Si](C)(C)NCCC(=O)O[Si](C)(C)C | 1272.7 | Standard non polar | 33892256 | | beta-Alanine,2TMS,isomer #1 | C[Si](C)(C)NCCC(=O)O[Si](C)(C)C | 1331.7 | Standard polar | 33892256 | | beta-Alanine,2TMS,isomer #2 | C[Si](C)(C)N(CCC(=O)O)[Si](C)(C)C | 1377.1 | Semi standard non polar | 33892256 | | beta-Alanine,2TMS,isomer #2 | C[Si](C)(C)N(CCC(=O)O)[Si](C)(C)C | 1349.2 | Standard non polar | 33892256 | | beta-Alanine,2TMS,isomer #2 | C[Si](C)(C)N(CCC(=O)O)[Si](C)(C)C | 1576.1 | Standard polar | 33892256 | | beta-Alanine,3TMS,isomer #1 | C[Si](C)(C)OC(=O)CCN([Si](C)(C)C)[Si](C)(C)C | 1446.2 | Semi standard non polar | 33892256 | | beta-Alanine,3TMS,isomer #1 | C[Si](C)(C)OC(=O)CCN([Si](C)(C)C)[Si](C)(C)C | 1413.5 | Standard non polar | 33892256 | | beta-Alanine,3TMS,isomer #1 | C[Si](C)(C)OC(=O)CCN([Si](C)(C)C)[Si](C)(C)C | 1368.0 | Standard polar | 33892256 | | beta-Alanine,1TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CCN | 1255.5 | Semi standard non polar | 33892256 | | beta-Alanine,1TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NCCC(=O)O | 1401.4 | Semi standard non polar | 33892256 | | beta-Alanine,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NCCC(=O)O[Si](C)(C)C(C)(C)C | 1658.1 | Semi standard non polar | 33892256 | | beta-Alanine,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NCCC(=O)O[Si](C)(C)C(C)(C)C | 1662.4 | Standard non polar | 33892256 | | beta-Alanine,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NCCC(=O)O[Si](C)(C)C(C)(C)C | 1611.9 | Standard polar | 33892256 | | beta-Alanine,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)N(CCC(=O)O)[Si](C)(C)C(C)(C)C | 1798.5 | Semi standard non polar | 33892256 | | beta-Alanine,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)N(CCC(=O)O)[Si](C)(C)C(C)(C)C | 1742.5 | Standard non polar | 33892256 | | beta-Alanine,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)N(CCC(=O)O)[Si](C)(C)C(C)(C)C | 1734.9 | Standard polar | 33892256 | | beta-Alanine,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CCN([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 2065.5 | Semi standard non polar | 33892256 | | beta-Alanine,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CCN([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 2025.4 | Standard non polar | 33892256 | | beta-Alanine,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CCN([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 1763.9 | Standard polar | 33892256 |
|
|---|
| Spectra |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-007k-1920000000-9389ee061bd6e9ae3b22 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-0fdt-2930000000-5799fddb7fac04d22372 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-006t-1940000000-49f1b7c80f35cbcf102e | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-006t-2920000000-c701f2e5652fa4f35e50 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-00di-8930000000-c4f8138d4cb7f1ea851a | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-MS (1 TMS) | splash10-014i-2900000000-c54f1d5878600393dca7 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-MS (2 TMS) | splash10-0udi-1900000000-2f55a09bba074034dee7 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-MS (3 TMS) | splash10-00ds-2940000000-4192453eaba2cbecf800 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine EI-B (Non-derivatized) | splash10-006t-1960000000-665868f9adaa275dbdb3 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-EI-TOF (Non-derivatized) | splash10-007k-1920000000-9389ee061bd6e9ae3b22 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-EI-TOF (Non-derivatized) | splash10-0fdt-2930000000-5799fddb7fac04d22372 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-EI-TOF (Non-derivatized) | splash10-006t-1940000000-49f1b7c80f35cbcf102e | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-EI-TOF (Non-derivatized) | splash10-006t-2920000000-c701f2e5652fa4f35e50 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-EI-TOF (Non-derivatized) | splash10-00di-8930000000-c4f8138d4cb7f1ea851a | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-MS (Non-derivatized) | splash10-00ds-2940000000-4192453eaba2cbecf800 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-MS (Non-derivatized) | splash10-014i-2900000000-c54f1d5878600393dca7 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-MS (Non-derivatized) | splash10-0udi-1900000000-2f55a09bba074034dee7 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-EI-TOF (Non-derivatized) | splash10-0udi-0900000000-efc0822e26bcf842e232 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - beta-Alanine GC-EI-TOF (Non-derivatized) | splash10-0072-2920000000-284cff207c712b6698d6 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - beta-Alanine GC-MS (Non-derivatized) - 70eV, Positive | splash10-001i-9000000000-ad7f96b2c6a1f5183794 | 2016-09-22 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - beta-Alanine GC-MS (1 TMS) - 70eV, Positive | splash10-0fk9-9700000000-d73baf312b8af27575b3 | 2017-10-06 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - beta-Alanine GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - beta-Alanine GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - beta-Alanine GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - beta-Alanine GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-001i-9000000000-be815b0a56f225da04d8 | 2014-09-20 | Not Available | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine Quattro_QQQ 10V, Positive-QTOF (Annotated) | splash10-00dl-9000000000-c30cc77c93e4ec09265a | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine Quattro_QQQ 25V, Positive-QTOF (Annotated) | splash10-000j-9000000000-540a52fcec629c0a0647 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine Quattro_QQQ 40V, Positive-QTOF (Annotated) | splash10-000b-9000000000-531b17f8e16896225186 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative-QTOF | splash10-000i-9000000000-61740a52186a51b8543c | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative-QTOF | splash10-000i-9000000000-1bdf967935160468d550 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative-QTOF | splash10-000i-9000000000-0ba5264cfc5ec108718b | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive-QTOF | splash10-0006-9000000000-c6a2c78d322328400127 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive-QTOF | splash10-00di-9000000000-d5f47100c42e7777802e | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive-QTOF | splash10-001i-9000000000-e8f43be75c153e5be3cc | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive-QTOF | splash10-1004-9000100000-ccb09493216d97165c8d | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive-QTOF | splash10-00l7-9230000000-644b1dbe52c5f07e285a | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive-QTOF | splash10-0006-9000000000-ff4a990aa1fb1b7cc33d | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - beta-Alanine LC-ESI-QQ , negative-QTOF | splash10-000i-9000000000-61740a52186a51b8543c | 2017-09-14 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - beta-Alanine 10V, Positive-QTOF | splash10-00di-9000000000-3ac278f3f0cfc3d50ebf | 2015-04-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - beta-Alanine 20V, Positive-QTOF | splash10-05fr-9000000000-6d6e140cf4317b214296 | 2015-04-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - beta-Alanine 40V, Positive-QTOF | splash10-05dl-9000000000-ead5967d7f21631b6fc6 | 2015-04-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - beta-Alanine 10V, Negative-QTOF | splash10-000i-9000000000-91312dd333b1413fa045 | 2015-04-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - beta-Alanine 20V, Negative-QTOF | splash10-007c-9000000000-caef7f2a75815a584747 | 2015-04-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - beta-Alanine 40V, Negative-QTOF | splash10-0fdo-9000000000-5642be49ea98b530ff09 | 2015-04-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - beta-Alanine 10V, Positive-QTOF | splash10-00di-9000000000-3ac278f3f0cfc3d50ebf | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - beta-Alanine 20V, Positive-QTOF | splash10-05fr-9000000000-6d6e140cf4317b214296 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - beta-Alanine 40V, Positive-QTOF | splash10-05dl-9000000000-ead5967d7f21631b6fc6 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - beta-Alanine 10V, Negative-QTOF | splash10-000i-9000000000-91312dd333b1413fa045 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - beta-Alanine 20V, Negative-QTOF | splash10-007c-9000000000-caef7f2a75815a584747 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - beta-Alanine 40V, Negative-QTOF | splash10-0fdo-9000000000-5642be49ea98b530ff09 | 2015-05-27 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Experimental 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | 2021-10-10 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
|
|---|
| Biological Properties |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Mitochondria
|
|---|
| Biospecimen Locations | - Blood
- Cerebrospinal Fluid (CSF)
- Feces
- Saliva
- Urine
|
|---|
| Tissue Locations | - Kidney
- Pancreas
- Placenta
- Prostate
- Skeletal Muscle
|
|---|
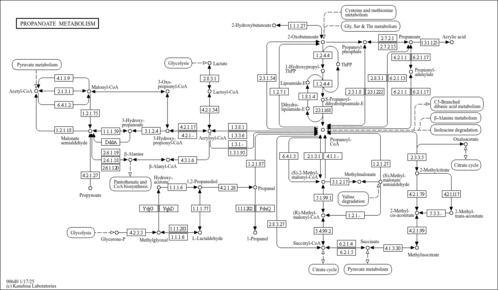
| Pathways | |
|---|
| Normal Concentrations |
|---|
| |
| Blood | Detected and Quantified | 3.8 +/- 2.9 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 1.76 +/- 0.74 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 3.0 +/- 0.85 uM | Adult (>18 years old) | Male | Normal | | details | | Blood | Detected and Quantified | 1.76 +/- 0.74 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 3.0 +/- 0.85 uM | Adult (>18 years old) | Male | Normal | | details | | Blood | Detected and Quantified | 3.0 (1.3-20.0) uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 1.76 +/- 0.74 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 3.00 +/- 0.85 uM | Adult (>18 years old) | Male | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 0.024 +/- 0.013 uM | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Not Specified | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Female | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Not Specified | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Saliva | Detected and Quantified | 1.01 +/- 0.448 uM | Adult (>18 years old) | Female | Normal | | details | | Saliva | Detected and Quantified | 1.48 +/- 0.851 uM | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected and Quantified | 1.14 +/- 1.34 uM | Adult (>18 years old) | Female | Normal | | details | | Saliva | Detected and Quantified | 1.02 +/- 1.25 uM | Adult (>18 years old) | Female | Normal | | details | | Saliva | Detected and Quantified | 2.71 +/- 0.74 uM | Adult (>18 years old) | Female | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 0.00068 +/- 0.00049 umol/mmol creatinine | Adult (>18 years old) | Male | Normal | | details | | Urine | Detected and Quantified | 7.24 (5.07-11.33) umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 0.23 +/- 0.73 umol/mmol creatinine | Infant (0-1 year old) | Both | Normal | | details | | Urine | Detected and Quantified | 0 +/- 5.65 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 10.18 +/- 10.18 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 24.87 +/- 62.18 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | <24.77 umol/mmol creatinine | Infant (0-1 year old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | <10.41 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | <2.83 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | <2.83 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | <5.54 umol/mmol creatinine | Adolescent (13-18 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | <5.88 umol/mmol creatinine | Adult (>18 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 0.0007 +/- 0.00075 umol/mmol creatinine | Adult (>18 years old) | Female | Normal | | details | | Urine | Detected and Quantified | 1.27 +/- 0.9 umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 0.8 umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 5.9 (3.4-13.0) umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 0.00068 +/- 0.00049 umol/mmol creatinine | Adult (>18 years old) | Male | Normal | | details | | Urine | Detected and Quantified | 0.0007 +/- 0.00075 umol/mmol creatinine | Adult (>18 years old) | Female | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 0.57-1.4 umol/mmol creatinine | Adult (>18 years old) | Male | Normal | | details | | Urine | Detected and Quantified | 0.58-1.1 umol/mmol creatinine | Adult (>18 years old) | Female | Normal | | details | | Urine | Detected and Quantified | 11.821 +/- 6.072 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 2.4 +/- 3.0 umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details |
|
|---|
| Abnormal Concentrations |
|---|
| |
| Blood | Detected and Quantified | 6 uM | Children (1-13 years old) | Male | Methylmalonate semialdehyde dehydrogenase deficiency | | details | | Blood | Detected and Quantified | 2.7 +/- 1.3 uM | Adult (>18 years old) | Both | Dihydropyrimidine dehydrogenase (DPD) deficiency | | details | | Blood | Detected and Quantified | 4.0 (0.9-6.2) uM | Adult (>18 years old) | Both | Dihydropyrimidine dehydrogenase (DPD) deficiency | | details | | Blood | Detected and Quantified | 23.0 uM | Children (1-13 years old) | Both | GABA transaminase deficiency | | details | | Blood | Detected and Quantified | 107.5 (15.00-200.00) uM | Children (1-13 years old) | Both | Hyper beta-alaninemia | | details | | Blood | Detected and Quantified | 5.00 (0.00-10.00) uM | Adult (>18 years old) | Both | Hyper beta-alaninemia | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 0.036 +/- 0.034 uM | Adult (>18 years old) | Both | Dihydropyrimidine dehydrogenase (DPD) deficiency | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 22.5 (0.00-45.00) uM | Children (1-13 years old) | Both | Hyper beta-alaninemia | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 0.48 uM | Children (1-13 years old) | Both | GABA transaminase deficiency | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 0.03 (0.00-0.06) uM | Adult (>18 years old) | Both | Gaba-transaminase deficiency | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Ulcerative colitis | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Crohn disease | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal Cancer | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Not Specified | Pancreatic cancer | | details | | Saliva | Detected and Quantified | 2.05 +/- 0.86 uM | Adult (>18 years old) | Both | Lewy body disease | | details | | Saliva | Detected and Quantified | 3.33 +/- 1.32 uM | Adult (>18 years old) | Male | Frontotemporal lobe dementia | | details | | Saliva | Detected and Quantified | 2.84 +/- 1.41 uM | Adult (>18 years old) | Male | Alzheimer's disease | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Not Specified | Periodontal diseases | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Female | Breast cancer | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Oral cancer | | details | | Urine | Detected and Quantified | 23.937 +/- 15.623 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Eosinophilic esophagitis | | details | | Urine | Detected and Quantified | 1.4 +/- 1.4 umol/mmol creatinine | Adult (>18 years old) | Both | Dihydropyrimidine dehydrogenase (DPD) deficiency | | details | | Urine | Detected and Quantified | 0.0-2.3 umol/mmol creatinine | Children (1-13 years old) | Both | GABA transaminase deficiency | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Bladder cancer | | details |
|
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | | Gaba-transaminase deficiency |
|---|
- G.Frauendienst-Egger, Friedrich K. Trefz (2017). MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de). METAGENE consortium.
| | Hyper beta-alaninemia |
|---|
- G.Frauendienst-Egger, Friedrich K. Trefz (2017). MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de). METAGENE consortium.
| | Methylmalonate semialdehyde dehydrogenase deficiency |
|---|
- Pollitt RJ, Green A, Smith R: Excessive excretion of beta-alanine and of 3-hydroxypropionic, R- and S-3-aminoisobutyric, R- and S-3-hydroxyisobutyric and S-2-(hydroxymethyl)butyric acids probably due to a defect in the metabolism of the corresponding malonic semialdehydes. J Inherit Metab Dis. 1985;8(2):75-9. [PubMed:3939535 ]
| | Colorectal cancer |
|---|
- Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP: Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016 Jun 6;4:11. doi: 10.1186/s40170-016-0151-y. eCollection 2016. [PubMed:27275383 ]
- Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ: Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016 Mar 25;11(3):e0152126. doi: 10.1371/journal.pone.0152126. eCollection 2016. [PubMed:27015276 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
| | Crohn's disease |
|---|
- Azario I, Pievani A, Del Priore F, Antolini L, Santi L, Corsi A, Cardinale L, Sawamoto K, Kubaski F, Gentner B, Bernardo ME, Valsecchi MG, Riminucci M, Tomatsu S, Aiuti A, Biondi A, Serafini M: Neonatal umbilical cord blood transplantation halts skeletal disease progression in the murine model of MPS-I. Sci Rep. 2017 Aug 25;7(1):9473. doi: 10.1038/s41598-017-09958-9. [PubMed:28842642 ]
| | Ulcerative colitis |
|---|
- Azario I, Pievani A, Del Priore F, Antolini L, Santi L, Corsi A, Cardinale L, Sawamoto K, Kubaski F, Gentner B, Bernardo ME, Valsecchi MG, Riminucci M, Tomatsu S, Aiuti A, Biondi A, Serafini M: Neonatal umbilical cord blood transplantation halts skeletal disease progression in the murine model of MPS-I. Sci Rep. 2017 Aug 25;7(1):9473. doi: 10.1038/s41598-017-09958-9. [PubMed:28842642 ]
| | Perillyl alcohol administration for cancer treatment |
|---|
- Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M: Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010 Mar;6(1):78-95. Epub 2009 Sep 10. [PubMed:20300169 ]
| | Pancreatic cancer |
|---|
- Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M: Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010 Mar;6(1):78-95. Epub 2009 Sep 10. [PubMed:20300169 ]
| | Periodontal disease |
|---|
- Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M: Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010 Mar;6(1):78-95. Epub 2009 Sep 10. [PubMed:20300169 ]
| | Alzheimer's disease |
|---|
- Tsuruoka M, Hara J, Hirayama A, Sugimoto M, Soga T, Shankle WR, Tomita M: Capillary electrophoresis-mass spectrometry-based metabolome analysis of serum and saliva from neurodegenerative dementia patients. Electrophoresis. 2013 Oct;34(19):2865-72. doi: 10.1002/elps.201300019. Epub 2013 Sep 6. [PubMed:23857558 ]
| | Frontotemporal dementia |
|---|
- Tsuruoka M, Hara J, Hirayama A, Sugimoto M, Soga T, Shankle WR, Tomita M: Capillary electrophoresis-mass spectrometry-based metabolome analysis of serum and saliva from neurodegenerative dementia patients. Electrophoresis. 2013 Oct;34(19):2865-72. doi: 10.1002/elps.201300019. Epub 2013 Sep 6. [PubMed:23857558 ]
| | Lewy body disease |
|---|
- Tsuruoka M, Hara J, Hirayama A, Sugimoto M, Soga T, Shankle WR, Tomita M: Capillary electrophoresis-mass spectrometry-based metabolome analysis of serum and saliva from neurodegenerative dementia patients. Electrophoresis. 2013 Oct;34(19):2865-72. doi: 10.1002/elps.201300019. Epub 2013 Sep 6. [PubMed:23857558 ]
| | Dihydropyrimidine dehydrogenase deficiency |
|---|
- Van Kuilenburg AB, Stroomer AE, Van Lenthe H, Abeling NG, Van Gennip AH: New insights in dihydropyrimidine dehydrogenase deficiency: a pivotal role for beta-aminoisobutyric acid? Biochem J. 2004 Apr 1;379(Pt 1):119-24. [PubMed:14705962 ]
| | Eosinophilic esophagitis |
|---|
- Slae, M., Huynh, H., Wishart, D.S. (2014). Analysis of 30 normal pediatric urine samples via NMR spectroscopy (unpublished work). NA.
|
|
|---|
| Associated OMIM IDs | - 613163 (Gaba-transaminase deficiency)
- 237400 (Hyper beta-alaninemia)
- 614105 (Methylmalonate semialdehyde dehydrogenase deficiency)
- 114500 (Colorectal cancer)
- 266600 (Crohn's disease)
- 260350 (Pancreatic cancer)
- 170650 (Periodontal disease)
- 104300 (Alzheimer's disease)
- 600274 (Frontotemporal dementia)
- 274270 (Dihydropyrimidine dehydrogenase deficiency)
- 610247 (Eosinophilic esophagitis)
|
|---|
| External Links |
|---|
| DrugBank ID | DB03107 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB002253 |
|---|
| KNApSAcK ID | C00001333 |
|---|
| Chemspider ID | 234 |
|---|
| KEGG Compound ID | C00099 |
|---|
| BioCyc ID | B-ALANINE |
|---|
| BiGG ID | 33848 |
|---|
| Wikipedia Link | Beta-Alanine |
|---|
| METLIN ID | 5119 |
|---|
| PubChem Compound | 239 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16958 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | ALA_B |
|---|
| MarkerDB ID | MDB00000028 |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Buc, Saul R.; Ford, Jared H.; Wise, E. C. Improved synthesis of b-alanine. Journal of the American Chemical Society (1945), 67 92-4. |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| General References | - van Kuilenburg AB, Meinsma R, Beke E, Assmann B, Ribes A, Lorente I, Busch R, Mayatepek E, Abeling NG, van Cruchten A, Stroomer AE, van Lenthe H, Zoetekouw L, Kulik W, Hoffmann GF, Voit T, Wevers RA, Rutsch F, van Gennip AH: beta-Ureidopropionase deficiency: an inborn error of pyrimidine degradation associated with neurological abnormalities. Hum Mol Genet. 2004 Nov 15;13(22):2793-801. Epub 2004 Sep 22. [PubMed:15385443 ]
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. doi: 10.1038/nature07762. [PubMed:19212411 ]
- Van Kuilenburg AB, Stroomer AE, Van Lenthe H, Abeling NG, Van Gennip AH: New insights in dihydropyrimidine dehydrogenase deficiency: a pivotal role for beta-aminoisobutyric acid? Biochem J. 2004 Apr 1;379(Pt 1):119-24. [PubMed:14705962 ]
- Malet-Martino MC, Bernadou J, Martino R, Armand JP: 19F NMR spectrometry evidence for bile acid conjugates of alpha-fluoro-beta-alanine as the main biliary metabolites of antineoplastic fluoropyrimidines in humans. Drug Metab Dispos. 1988 Jan-Feb;16(1):78-84. [PubMed:2894959 ]
- Klebanov GI, Teselkin YuO, Babenkova IV, Lyubitsky OB, Rebrova OYu, Boldyrev AA, Vladimirov YuA: Effect of carnosine and its components on free-radical reactions. Membr Cell Biol. 1998;12(1):89-99. [PubMed:9829262 ]
- Aznar J, Gilabert J, Estelles A, Fernandez MA, Villa P, Aznar JA: Evaluation of the soluble fibrin monomer complexes and other coagulation parameters in obstetric patients. Thromb Res. 1982 Sep 15;27(6):691-701. [PubMed:7179210 ]
- Champion EE, Mann SJ, Glazier JD, Jones CJ, Rawlings JM, Sibley CP, Greenwood SL: System beta and system A amino acid transporters in the feline endotheliochorial placenta. Am J Physiol Regul Integr Comp Physiol. 2004 Dec;287(6):R1369-79. Epub 2004 Jul 29. [PubMed:15284084 ]
- Kuo KC, Cole TF, Gehrke CW, Waalkes TP, Borek E: Dual-column cation-exchange chromatographic method for beta-aminoisobutyric acid and beta-alanine in biological samples. Clin Chem. 1978 Aug;24(8):1373-80. [PubMed:679461 ]
- Heggie GD, Sommadossi JP, Cross DS, Huster WJ, Diasio RB: Clinical pharmacokinetics of 5-fluorouracil and its metabolites in plasma, urine, and bile. Cancer Res. 1987 Apr 15;47(8):2203-6. [PubMed:3829006 ]
- Gibson KM, Schor DS, Gupta M, Guerand WS, Senephansiri H, Burlingame TG, Bartels H, Hogema BM, Bottiglieri T, Froestl W, Snead OC, Grompe M, Jakobs C: Focal neurometabolic alterations in mice deficient for succinate semialdehyde dehydrogenase. J Neurochem. 2002 Apr;81(1):71-9. [PubMed:12067239 ]
- Holm B, Nilsen DW, Kierulf P, Godal HC: Purification and characterization of 3 fibrinogens with different molecular weights obtained from normal human plasma. Thromb Res. 1985 Jan 1;37(1):165-76. [PubMed:3983897 ]
- Chen Y, Getchell TV, Sparks DL, Getchell ML: Cellular localization of carnosinase in the human nasal mucosa. Acta Otolaryngol. 1994 Mar;114(2):193-8. [PubMed:8203202 ]
- Milasta S, Pediani J, Appelbe S, Trim S, Wyatt M, Cox P, Fidock M, Milligan G: Interactions between the Mas-related receptors MrgD and MrgE alter signalling and trafficking of MrgD. Mol Pharmacol. 2006 Feb;69(2):479-91. Epub 2005 Nov 9. [PubMed:16282220 ]
- Harris RC, Tallon MJ, Dunnett M, Boobis L, Coakley J, Kim HJ, Fallowfield JL, Hill CA, Sale C, Wise JA: The absorption of orally supplied beta-alanine and its effect on muscle carnosine synthesis in human vastus lateralis. Amino Acids. 2006 May;30(3):279-89. Epub 2006 Mar 24. [PubMed:16554972 ]
- Hibbard JU, Pridjian G, Whitington PF, Moawad AH: Taurine transport in the in vitro perfused human placenta. Pediatr Res. 1990 Jan;27(1):80-4. [PubMed:2296474 ]
- Karmanskii IM: [Effect of pepsin on low density serum lipoproteins]. Vopr Med Khim. 1977 Jul-Aug;23(4):530-4. [PubMed:200005 ]
- Johnson MR, Barnes S, Sweeny DJ, Diasio RB: 2-Fluoro-beta-alanine, a previously unrecognized substrate for bile acid coenzyme A:amino acid:N-acyltransferase from human liver. Biochem Pharmacol. 1990 Sep 15;40(6):1241-6. [PubMed:2119585 ]
- Shetewy A, Shimada-Takaura K, Warner D, Jong CJ, Mehdi AB, Alexeyev M, Takahashi K, Schaffer SW: Mitochondrial defects associated with beta-alanine toxicity: relevance to hyper-beta-alaninemia. Mol Cell Biochem. 2016 May;416(1-2):11-22. doi: 10.1007/s11010-016-2688-z. Epub 2016 Mar 29. [PubMed:27023909 ]
- Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. [PubMed:32033212 ]
|
|---|