| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2023-02-21 17:15:04 UTC |
|---|
| HMDB ID | HMDB0000700 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Hydroxypropionic acid |
|---|
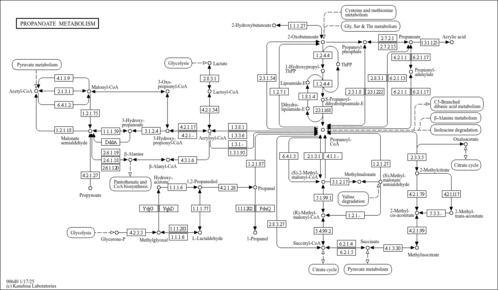
| Description | 3-Hydroxypropionic acid is a carboxylic acid. It is an intermediate in the breakdown of branched-chain amino acids and propionic acid from the gut. Typically it originates from propionyl-CoA and a defect in the enzyme propionyl carboxylase. This leads to a buildup in propionyl-CoA in the mitochondria. Such a buildup can lead to a disruption of the esterified CoA:free CoA ratio and ultimately to mitochondrial toxicity. Detoxification of these metabolic end products occurs via the transfer of the propionyl moiety to carnitine-forming propionyl-carnitine, which is then transferred across the inner mitochondrial membrane. 3-Hydroxypropionic acid is then released as the free acid. As an industrial chemical, it is used in the production of various chemicals such as acrylates in industry. When present in sufficiently high levels, 3-hydroxypropionic acid can act as an acidogen and a metabotoxin. An acidogen is an acidic compound that induces acidosis, which has multiple adverse effects on many organ systems. A metabotoxin is an endogenously produced metabolite that causes adverse health effects at chronically high levels. Chronically high levels of hydroxypropionic acid are associated with many inborn errors of metabolism including biotinidase deficiency, malonic aciduria, methylmalonate semialdehyde dehydrogenase deficiency, methylmalonic aciduria, methylmalonic aciduria due to cobalamin-related disorders, and propionic acidemia. Hydroxypropionic acid is an organic acid. Abnormally high levels of organic acids in the blood (organic acidemia), urine (organic aciduria), the brain, and other tissues lead to general metabolic acidosis. Acidosis typically occurs when arterial pH falls below 7.35. Infants with acidosis have symptoms that include poor feeding, vomiting, loss of appetite, weak muscle tone (hypotonia), and lack of energy (lethargy). These can progress to heart, liver, and kidney abnormalities, seizures, coma, and possibly death. These are also the characteristic symptoms of the IEMs mentioned above. Many affected children with organic acidemias experience intellectual disability or delayed development. In adults, acidosis or acidemia is characterized by headaches, confusion, feeling tired, tremors, sleepiness, and seizures. 3-Hydroxypropionic acid is also a microbial metabolite found in Escherichia, Klebsiella and Saccharomyces (PMID: 26360870 ). |
|---|
| Structure | InChI=1S/C3H6O3/c4-2-1-3(5)6/h4H,1-2H2,(H,5,6) |
|---|
| Synonyms | | Value | Source |
|---|
| 3-HYDROXY-propanoIC ACID | ChEBI | | 3-Hydroxypropanoic acid | ChEBI | | beta-Hydroxypropionic acid | ChEBI | | Hydracrylic acid | ChEBI | | 3-Hydroxypropionate | Kegg | | 3-Hydroxypropionic acid | Kegg | | 3-HYDROXY-propanoate | Generator | | 3-Hydroxypropanoate | Generator | | b-Hydroxypropionate | Generator | | b-Hydroxypropionic acid | Generator | | beta-Hydroxypropionate | Generator | | Β-hydroxypropionate | Generator | | Β-hydroxypropionic acid | Generator | | Hydracrylate | Generator | | Hydroxypropionate | Generator | | 2-Deoxyglycerate | HMDB | | 2-Deoxyglyceric acid | HMDB | | b-Lactate | HMDB | | b-Lactic acid | HMDB | | beta-Lactate | HMDB | | beta-Lactic acid | HMDB | | Ethylenelactate | HMDB | | Ethylenelactic acid | HMDB |
|
|---|
| Chemical Formula | C3H6O3 |
|---|
| Average Molecular Weight | 90.0779 |
|---|
| Monoisotopic Molecular Weight | 90.031694058 |
|---|
| IUPAC Name | 3-hydroxypropanoic acid |
|---|
| Traditional Name | hydroxypropionic acid |
|---|
| CAS Registry Number | 503-66-2 |
|---|
| SMILES | OCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C3H6O3/c4-2-1-3(5)6/h4H,1-2H2,(H,5,6) |
|---|
| InChI Key | ALRHLSYJTWAHJZ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as beta hydroxy acids and derivatives. Beta hydroxy acids and derivatives are compounds containing a carboxylic acid substituted with a hydroxyl group on the C3 carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Hydroxy acids and derivatives |
|---|
| Sub Class | Beta hydroxy acids and derivatives |
|---|
| Direct Parent | Beta hydroxy acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Beta-hydroxy acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Primary alcohol
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | < 25 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 1.33 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 8.7119 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 5.11 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 257.5 seconds | 40023050 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 665.4 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 343.3 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 67.6 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 238.3 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 71.6 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 262.9 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 250.1 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 476.1 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 568.9 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 47.5 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 770.2 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 215.5 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 265.3 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 680.5 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 301.4 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 343.2 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Hydroxypropionic acid,1TMS,isomer #1 | C[Si](C)(C)OCCC(=O)O | 1077.3 | Semi standard non polar | 33892256 | | Hydroxypropionic acid,1TMS,isomer #2 | C[Si](C)(C)OC(=O)CCO | 992.0 | Semi standard non polar | 33892256 | | Hydroxypropionic acid,2TMS,isomer #1 | C[Si](C)(C)OCCC(=O)O[Si](C)(C)C | 1155.0 | Semi standard non polar | 33892256 | | Hydroxypropionic acid,1TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OCCC(=O)O | 1314.8 | Semi standard non polar | 33892256 | | Hydroxypropionic acid,1TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)OC(=O)CCO | 1220.4 | Semi standard non polar | 33892256 | | Hydroxypropionic acid,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OCCC(=O)O[Si](C)(C)C(C)(C)C | 1567.0 | Semi standard non polar | 33892256 |
|
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - Hydroxypropionic acid GC-EI-TOF (Non-derivatized) | splash10-0002-0900000000-26c10a7c09d2819f18b3 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Hydroxypropionic acid GC-EI-TOF (Non-derivatized) | splash10-0fba-0900000000-8bf04c0af8fd2e3b58de | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Hydroxypropionic acid GC-EI-TOF (Non-derivatized) | splash10-0002-0900000000-26c10a7c09d2819f18b3 | 2018-05-18 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Hydroxypropionic acid GC-EI-TOF (Non-derivatized) | splash10-0fba-0900000000-8bf04c0af8fd2e3b58de | 2018-05-18 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Hydroxypropionic acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-0007-9000000000-9aeb0d9c81f19a27c424 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Hydroxypropionic acid GC-MS (2 TMS) - 70eV, Positive | splash10-0100-9720000000-bbec0186b2d36c22228d | 2017-10-06 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Hydroxypropionic acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Hydroxypropionic acid GC-MS (TMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Hydroxypropionic acid GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Hydroxypropionic acid GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Hydroxypropionic acid GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Hydroxypropionic acid GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid Quattro_QQQ 10V, Positive-QTOF (Annotated) | splash10-00di-9000000000-3f5b72b8462bfcb4f391 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid Quattro_QQQ 25V, Positive-QTOF (Annotated) | splash10-00di-9000000000-3e9fa999dd5a46584db4 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid Quattro_QQQ 40V, Positive-QTOF (Annotated) | splash10-03fu-5900000000-bd7d31d2678f47785461 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative-QTOF | splash10-000i-9000000000-d440d21b0f161a9853f1 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative-QTOF | splash10-0a4r-9000000000-242c99f8bca595fe224e | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative-QTOF | splash10-052f-9000000000-a9bac0e4116df119abb1 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative-QTOF | splash10-0006-9000000000-081d862d9b4bc28ae21b | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative-QTOF | splash10-0006-9000000000-be05ae87088e43aa060d | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid LC-ESI-QQ , negative-QTOF | splash10-000i-9000000000-d440d21b0f161a9853f1 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid LC-ESI-QQ , negative-QTOF | splash10-0a4r-9000000000-dba7e70fabc7dac6fe4b | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid LC-ESI-QQ , negative-QTOF | splash10-052f-9000000000-a9bac0e4116df119abb1 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid LC-ESI-QQ , negative-QTOF | splash10-0006-9000000000-081d862d9b4bc28ae21b | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid LC-ESI-QQ , negative-QTOF | splash10-0006-9000000000-be05ae87088e43aa060d | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid 20V, Positive-QTOF | splash10-01ox-9000000000-abdb98d8c1686dc43cc3 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid 40V, Positive-QTOF | splash10-03di-9000000000-7e9710b33d0c0f60adc0 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid 20V, Positive-QTOF | splash10-00kf-9000000000-8076ca0477f5e951c57d | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid 10V, Positive-QTOF | splash10-03dl-9000000000-74725037ead28bc3cbb5 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid 40V, Positive-QTOF | splash10-029l-9000000000-e17da766d94d48429d67 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Hydroxypropionic acid 10V, Positive-QTOF | splash10-03di-9000000000-e19c639a151e1fca0d34 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hydroxypropionic acid 10V, Positive-QTOF | splash10-00di-9000000000-9e7369e67b8849650d52 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hydroxypropionic acid 20V, Positive-QTOF | splash10-05fs-9000000000-6bc03f735cf4358adaaf | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hydroxypropionic acid 40V, Positive-QTOF | splash10-05di-9000000000-8c197b7691bbbacb263a | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hydroxypropionic acid 10V, Negative-QTOF | splash10-000i-9000000000-1983baaf50a36f58c36c | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hydroxypropionic acid 20V, Negative-QTOF | splash10-00dr-9000000000-5cc8fd5907f4674c72f3 | 2015-05-27 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hydroxypropionic acid 40V, Negative-QTOF | splash10-006x-9000000000-3e0c27682d8f7902d021 | 2015-05-27 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
|
|---|
| Disease References | | Stomach cancer |
|---|
- Ikeda A, Nishiumi S, Shinohara M, Yoshie T, Hatano N, Okuno T, Bamba T, Fukusaki E, Takenawa T, Azuma T, Yoshida M: Serum metabolomics as a novel diagnostic approach for gastrointestinal cancer. Biomed Chromatogr. 2012 May;26(5):548-58. doi: 10.1002/bmc.1671. Epub 2011 Jul 20. [PubMed:21773981 ]
| | Branched-chain Keto Acid Dehydrogenase Kinase Deficiency |
|---|
- Novarino G, El-Fishawy P, Kayserili H, Meguid NA, Scott EM, Schroth J, Silhavy JL, Kara M, Khalil RO, Ben-Omran T, Ercan-Sencicek AG, Hashish AF, Sanders SJ, Gupta AR, Hashem HS, Matern D, Gabriel S, Sweetman L, Rahimi Y, Harris RA, State MW, Gleeson JG: Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science. 2012 Oct 19;338(6105):394-7. doi: 10.1126/science.1224631. Epub 2012 Sep 6. [PubMed:22956686 ]
| | Colorectal cancer |
|---|
- Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP: Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016 Jun 6;4:11. doi: 10.1186/s40170-016-0151-y. eCollection 2016. [PubMed:27275383 ]
- Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ: Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016 Mar 25;11(3):e0152126. doi: 10.1371/journal.pone.0152126. eCollection 2016. [PubMed:27015276 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
| | Biotinidase deficiency |
|---|
- G.Frauendienst-Egger, Friedrich K. Trefz (2017). MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de). METAGENE consortium.
| | Eosinophilic esophagitis |
|---|
- Slae, M., Huynh, H., Wishart, D.S. (2014). Analysis of 30 normal pediatric urine samples via NMR spectroscopy (unpublished work). NA.
| | Propionic acidemia |
|---|
- Riemersma M, Hazebroek MR, Helderman-van den Enden ATJM, Salomons GS, Ferdinandusse S, Brouwers MCGJ, van der Ploeg L, Heymans S, Glatz JFC, van den Wijngaard A, Krapels IPC, Bierau J, Brunner HG: Propionic acidemia as a cause of adult-onset dilated cardiomyopathy. Eur J Hum Genet. 2017 Nov;25(11):1195-1201. doi: 10.1038/ejhg.2017.127. Epub 2017 Aug 30. [PubMed:28853722 ]
- G.Frauendienst-Egger, Friedrich K. Trefz (2017). MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de). METAGENE consortium.
|
|
|---|