| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2023-02-21 17:14:53 UTC |
|---|
| HMDB ID | HMDB0000500 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 4-Hydroxybenzoic acid |
|---|
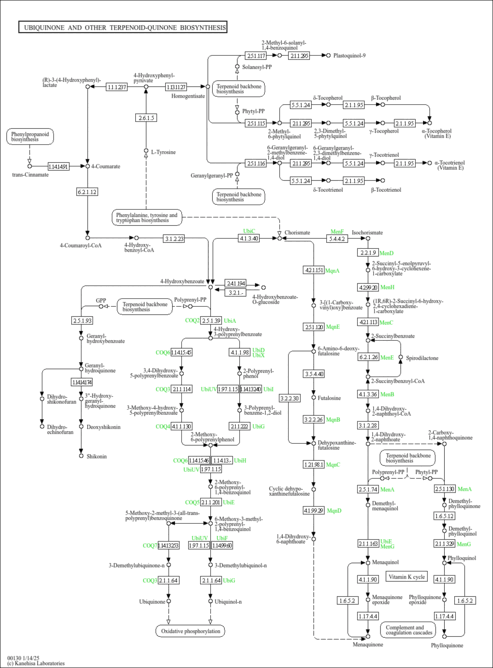
| Description | 4-Hydroxybenzoic acid, also known as p-hydroxybenzoate or 4-carboxyphenol, belongs to the class of organic compounds known as hydroxybenzoic acid derivatives. Hydroxybenzoic acid derivatives are compounds containing a hydroxybenzoic acid (or a derivative), which is a benzene ring bearing a carboxyl and a hydroxyl groups. 4-Hydroxybenzoic acid is a white crystalline solid that is slightly soluble in water and chloroform but more soluble in polar organic solvents such as alcohols and acetone. It is a nutty and phenolic tasting compound. 4-Hydroxybenzoic acid exists in all living species, ranging from bacteria to plants to humans. 4-Hydroxybenzoic acid can be found naturally in coconut. It is one of the main catechins metabolites found in humans after consumption of green tea infusions. It is also found in wine, in vanilla, in Açaí oil, obtained from the fruit of the açaí palm (Euterpe oleracea), at relatively high concetrations (892±52 mg/kg). It is also found in cloudy olive oil and in the edible mushroom Russula virescens. It has been detected in red huckleberries, rabbiteye blueberries, and corianders and in a lower concentration in olives, red raspberries, and almonds. In humans, 4-hydroxybenzoic acid is involved in ubiquinone biosynthesis. In particular, the enzyme 4-hydroxybenzoate polyprenyltransferase uses a polyprenyl diphosphate and 4-hydroxybenzoate to produce diphosphate and 4-hydroxy-3-polyprenylbenzoate. This enzyme participates in ubiquinone biosynthesis. 4-Hydroxybenzoic acid can be biosynthesized by the enzyme Chorismate lyase. Chorismate lyase is an enzyme that transforms chorismate into 4-hydroxybenzoate and pyruvate. This enzyme catalyses the first step in ubiquinone biosynthesis in Escherichia coli and other Gram-negative bacteria. 4-Hydroxybenzoate is an intermediate in many enzyme-mediated reactions in microbes. For instance, the enzyme 4-hydroxybenzaldehyde dehydrogenase uses 4-hydroxybenzaldehyde, NAD+ and H2O to produce 4-hydroxybenzoate, NADH and H+. This enzyme participates in toluene and xylene degradation in bacteria such as Pseudomonas mendocina. 4-hydroxybenzaldehyde dehydrogenase is also found in carrots. The enzyme 4-hydroxybenzoate 1-hydroxylase transforms 4-hydroxybenzoate, NAD(P)H, 2 H+ and O2 into hydroquinone, NAD(P)+, H2O and CO2. This enzyme participates in 2,4-dichlorobenzoate degradation and is found in Candida parapsilosis. The enzyme 4-hydroxybenzoate 3-monooxygenase transforms 4-hydroxybenzoate, NADPH, H+ and O2 into protocatechuate, NADP+ and H2O. This enzyme participates in benzoate degradation via hydroxylation and 2,4-dichlorobenzoate degradation and is found in Pseudomonas putida and Pseudomonas fluorescens. 4-Hydroxybenzoic acid is a popular antioxidant in part because of its low toxicity. 4-Hydroxybenzoic acid has estrogenic activity both in vitro and in vivo (PMID 9417843 ). |
|---|
| Structure | InChI=1S/C7H6O3/c8-6-3-1-5(2-4-6)7(9)10/h1-4,8H,(H,9,10) |
|---|
| Synonyms | | Value | Source |
|---|
| 4-Carboxyphenol | ChEBI | | p-HYDROXYBENZOIC ACID | ChEBI | | p-Salicylic acid | ChEBI | | Hydroxybenzoic acid | Kegg | | Hydroxybenzenecarboxylic acid | Kegg | | p-HYDROXYBENZOate | Generator | | p-Salicylate | Generator | | Hydroxybenzoate | Generator | | Hydroxybenzenecarboxylate | Generator | | 4-Hydroxybenzoate | Generator | | 4-Hydroxy-benzoate | HMDB | | 4-Hydroxy-benzoesaeure | HMDB | | 4-Hydroxy-benzoic acid | HMDB | | p-Carboxyphenol | HMDB | | p-Hydroxy-benzoate | HMDB | | p-Hydroxy-benzoic acid | HMDB | | Paraben-acid | HMDB | | 4-Hydroxybenzoic acid, calcium salt | HMDB | | 4-Hydroxybenzoic acid, dilithium salt | HMDB | | 4-Hydroxybenzoic acid, disodium salt | HMDB | | Para-hydroxybenzoic acid | HMDB | | Sodium p-hydroxybenzoate tetrahydrate | HMDB | | 4-Hydroxybenzoic acid, copper(2+)(1:1) salt | HMDB | | 4-Hydroxybenzoic acid, dipotassium salt | HMDB | | 4-Hydroxybenzoic acid, monopotassium salt | HMDB | | 4-Hydroxybenzoic acid, monosodium salt | HMDB | | 4-Hydroxybenzoic acid, monosodium salt, 11C-labeled | HMDB | | 4-Hydroxybenzene carboxylic acid | HMDB | | p-Hydroxyl benzoic acid | HMDB | | 4-Hydroxybenzoic acid | HMDB |
|
|---|
| Chemical Formula | C7H6O3 |
|---|
| Average Molecular Weight | 138.122 |
|---|
| Monoisotopic Molecular Weight | 138.031694053 |
|---|
| IUPAC Name | 4-hydroxybenzoic acid |
|---|
| Traditional Name | P-hydroxybenzoic acid |
|---|
| CAS Registry Number | 99-96-7 |
|---|
| SMILES | OC(=O)C1=CC=C(O)C=C1 |
|---|
| InChI Identifier | InChI=1S/C7H6O3/c8-6-3-1-5(2-4-6)7(9)10/h1-4,8H,(H,9,10) |
|---|
| InChI Key | FJKROLUGYXJWQN-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as hydroxybenzoic acid derivatives. Hydroxybenzoic acid derivatives are compounds containing a hydroxybenzoic acid (or a derivative), which is a benzene ring bearing a carboxyl and a hydroxyl groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Benzene and substituted derivatives |
|---|
| Sub Class | Benzoic acids and derivatives |
|---|
| Direct Parent | Hydroxybenzoic acid derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hydroxybenzoic acid
- Benzoic acid
- Benzoyl
- 1-hydroxy-2-unsubstituted benzenoid
- Phenol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | |
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 3.02 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 9.5686 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 3.53 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 71.1 seconds | 40023050 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 1089.7 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 331.3 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 101.1 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 205.9 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 112.0 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 341.8 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 337.6 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 112.2 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 706.5 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 275.6 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 952.9 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 227.4 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 288.4 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 505.2 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 230.3 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 201.4 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| 4-Hydroxybenzoic acid,1TMS,isomer #1 | C[Si](C)(C)OC(=O)C1=CC=C(O)C=C1 | 1590.9 | Semi standard non polar | 33892256 | | 4-Hydroxybenzoic acid,1TMS,isomer #2 | C[Si](C)(C)OC1=CC=C(C(=O)O)C=C1 | 1623.4 | Semi standard non polar | 33892256 | | 4-Hydroxybenzoic acid,2TMS,isomer #1 | C[Si](C)(C)OC(=O)C1=CC=C(O[Si](C)(C)C)C=C1 | 1642.4 | Semi standard non polar | 33892256 | | 4-Hydroxybenzoic acid,1TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)C1=CC=C(O)C=C1 | 1848.1 | Semi standard non polar | 33892256 | | 4-Hydroxybenzoic acid,1TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)OC1=CC=C(C(=O)O)C=C1 | 1867.2 | Semi standard non polar | 33892256 | | 4-Hydroxybenzoic acid,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)C1=CC=C(O[Si](C)(C)C(C)(C)C)C=C1 | 2141.0 | Semi standard non polar | 33892256 |
|
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (2 TMS) | splash10-01b9-0490000000-89473836b0071542185e | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-00xu-2890000000-dd5367ba838ccd5b29d8 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (2 TMS) | splash10-00di-9540000000-11db590137f79b7a32bf | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid GC-MS (2 TMS) | splash10-00xu-3890000000-7e553522b6ec5c075e25 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid EI-B (Non-derivatized) | splash10-00dr-5900000000-9b1d88421f1e1ded16f9 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid EI-B (Non-derivatized) | splash10-00di-9700000000-a9bb4b71d8b23a56ca67 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid EI-B (Non-derivatized) | splash10-0079-6900000000-617d58eb059c1e918ca0 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid EI-B (Non-derivatized) | splash10-00dr-4900000000-97bfa38b0d6bc152d470 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid EI-B (Non-derivatized) | splash10-00dr-7900000000-290af590b1fc44a86969 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid EI-B (Non-derivatized) | splash10-01b9-0490000000-d6001fb50397705d3e0b | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid GC-EI-TOF (Non-derivatized) | splash10-01b9-0490000000-89473836b0071542185e | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid GC-EI-TOF (Non-derivatized) | splash10-00xu-2890000000-dd5367ba838ccd5b29d8 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid GC-EI-TOF (Non-derivatized) | splash10-00di-9540000000-11db590137f79b7a32bf | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid GC-MS (Non-derivatized) | splash10-00xu-3890000000-7e553522b6ec5c075e25 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 4-Hydroxybenzoic acid GC-EI-TOF (Non-derivatized) | splash10-00xu-2980000000-0ba0ea7d63bd7fff3b22 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 4-Hydroxybenzoic acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-0079-5900000000-fcde26d72ca8445dff7e | 2016-09-22 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 4-Hydroxybenzoic acid GC-MS (2 TMS) - 70eV, Positive | splash10-00xu-7950000000-cc553d409cc2a872749d | 2017-10-06 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 4-Hydroxybenzoic acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 4-Hydroxybenzoic acid GC-MS (TMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 4-Hydroxybenzoic acid GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 4-Hydroxybenzoic acid GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 4-Hydroxybenzoic acid GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 4-Hydroxybenzoic acid GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-00dr-6900000000-71548845dbf99346758d | 2015-03-01 | Not Available | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid Quattro_QQQ 10V, Negative-QTOF (Annotated) | splash10-000f-9400000000-9fb9f8fbd7cf90a88604 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid Quattro_QQQ 25V, Negative-QTOF (Annotated) | splash10-0006-9000000000-f5d09d09184f72ec3bd7 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid Quattro_QQQ 40V, Negative-QTOF (Annotated) | splash10-0006-9000000000-d31cea608d0764edf5d0 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid EI-B (HITACHI M-68) , Positive-QTOF | splash10-00dr-5900000000-9b1d88421f1e1ded16f9 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid EI-B (HITACHI M-60) , Positive-QTOF | splash10-00di-9700000000-3ac766249fee4c68b8f4 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid EI-B (HITACHI M-80) , Positive-QTOF | splash10-0079-6900000000-4e15a4102e0b3bc8ddce | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative-QTOF | splash10-0a4r-9800000000-9e7208bfb79f9cca336b | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative-QTOF | splash10-0a4i-9000000000-1ed6d8123d44d126ed64 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative-QTOF | splash10-0a4i-9000000000-63a0e9c65e315bb89281 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative-QTOF | splash10-0a4i-9000000000-a8befcaa45eced4a9a51 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative-QTOF | splash10-0a4i-9000000000-26788d399f2b459abc21 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive-QTOF | splash10-0072-6900000000-ba06f5c422d59cd10cea | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive-QTOF | splash10-0002-9100000000-193fd4eb0cf324fb3abc | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative-QTOF | splash10-0006-9200000000-c834f7be13341f3e19b8 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid DI-ESI-qTof , Positive-QTOF | splash10-00di-0900000000-4f58e660923e896589a3 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid LC-ESI-QQ , negative-QTOF | splash10-0a4r-9800000000-9e7208bfb79f9cca336b | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid LC-ESI-QQ , negative-QTOF | splash10-0a4i-9000000000-1ed6d8123d44d126ed64 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid LC-ESI-QQ , negative-QTOF | splash10-0a4i-9000000000-e80d9637393e63e2d07c | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 4-Hydroxybenzoic acid LC-ESI-QQ , negative-QTOF | splash10-0a4i-9000000000-a8befcaa45eced4a9a51 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 4-Hydroxybenzoic acid 10V, Positive-QTOF | splash10-0079-0900000000-6351b89d53d1742a1fbb | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 4-Hydroxybenzoic acid 20V, Positive-QTOF | splash10-00di-1900000000-bdd873f3d1dc0f2af556 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 4-Hydroxybenzoic acid 40V, Positive-QTOF | splash10-00fu-9400000000-37262c1edda5dec21a19 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 4-Hydroxybenzoic acid 10V, Negative-QTOF | splash10-000i-3900000000-fcd512c9827982e7c9aa | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 4-Hydroxybenzoic acid 20V, Negative-QTOF | splash10-000f-9600000000-36013030005316a50fab | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 4-Hydroxybenzoic acid 40V, Negative-QTOF | splash10-0006-9100000000-5efa05442f95e2900559 | 2016-09-12 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
|
|---|
| Disease References | | Supradiaphragmatic malignancy |
|---|
- Goudas LC, Langlade A, Serrie A, Matson W, Milbury P, Thurel C, Sandouk P, Carr DB: Acute decreases in cerebrospinal fluid glutathione levels after intracerebroventricular morphine for cancer pain. Anesth Analg. 1999 Nov;89(5):1209-15. [PubMed:10553836 ]
| | Colorectal cancer |
|---|
- Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ: Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016 Mar 25;11(3):e0152126. doi: 10.1371/journal.pone.0152126. eCollection 2016. [PubMed:27015276 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
| | Eosinophilic esophagitis |
|---|
- Slae, M., Huynh, H., Wishart, D.S. (2014). Analysis of 30 normal pediatric urine samples via NMR spectroscopy (unpublished work). NA.
|
|
|---|
| General References | - Guneral F, Bachmann C: Age-related reference values for urinary organic acids in a healthy Turkish pediatric population. Clin Chem. 1994 Jun;40(6):862-6. [PubMed:8087979 ]
- Soni MG, Taylor SL, Greenberg NA, Burdock GA: Evaluation of the health aspects of methyl paraben: a review of the published literature. Food Chem Toxicol. 2002 Oct;40(10):1335-73. [PubMed:12387298 ]
- Shibusawa H, Sano Y, Yamamoto T, Kambegawa A, Ohkawa T, Satoh N, Okinaga S, Arai K: A radioimmunoassay of serum 16alpha-hydroxypregnenolone with specific antiserum. Endocrinol Jpn. 1978 Apr;25(2):185-9. [PubMed:668632 ]
- Wiebe LI, Mercer JR, Ryan AJ: Urinary metabolites of 3,5-di-(1-[13C]methyl-1-methylethyl)4-hydroxytoluene (BHT-13C) in man. Drug Metab Dispos. 1978 May-Jun;6(3):296-302. [PubMed:26551 ]
- Sakurai M, Ohsako M, Nagano M, Nakamura C, Tsuzuki O, Ichikawa M, Matsumoto Y: [Effect of human serum albumin on transport of drugs through human erythrocyte membranes]. Yakugaku Zasshi. 1996 Aug;116(8):630-8. [PubMed:8831264 ]
- Soni MG, Burdock GA, Taylor SL, Greenberg NA: Safety assessment of propyl paraben: a review of the published literature. Food Chem Toxicol. 2001 Jun;39(6):513-32. [PubMed:11346481 ]
- Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K: Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. 2018 Feb;57(1):1-24. doi: 10.1007/s00394-017-1445-8. Epub 2017 Apr 9. [PubMed:28393285 ]
- Peng X, Adachi K, Chen C, Kasai H, Kanoh K, Shizuri Y, Misawa N: Discovery of a marine bacterium producing 4-hydroxybenzoate and its alkyl esters, parabens. Appl Environ Microbiol. 2006 Aug;72(8):5556-61. doi: 10.1128/AEM.00494-06. [PubMed:16885309 ]
- Kallscheuer N, Marienhagen J: Corynebacterium glutamicum as platform for the production of hydroxybenzoic acids. Microb Cell Fact. 2018 May 12;17(1):70. doi: 10.1186/s12934-018-0923-x. [PubMed:29753327 ]
- Koistinen VM (2019). Effects of Food Processing and Gut Microbial Metabolism on Whole Grain Phytochemicals: A Metabolomics Approach. In Publications of the University of Eastern Finland. Dissertations in Health Sciences., no 510 (pp. 26-58). University of Eastern Finland. [ISBN:978-952-61-3088-0 ]
|
|---|