| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:49:08 UTC |
|---|
| HMDB ID | HMDB0001088 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Butyryl-CoA |
|---|
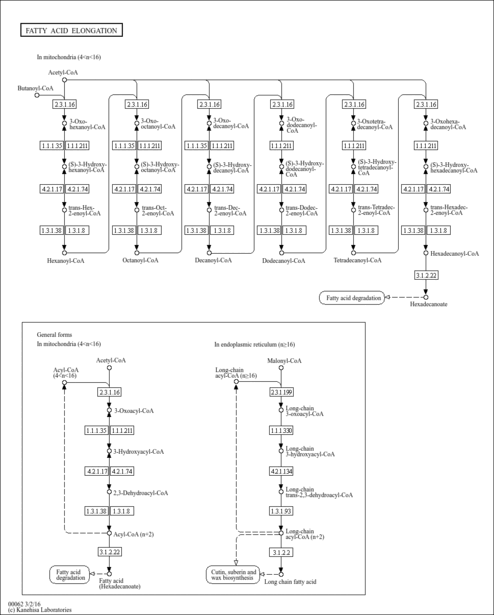
| Description | Butyryl-CoA is an intermediate in the metabolism of Butanoate. It is a substrate for Acyl-coenzyme A oxidase 3 (peroxisomal), 3-ketoacyl-CoA thiolase (mitochondrial), 3-ketoacyl-CoA thiolase (peroxisomal), Acyl-coenzyme A oxidase 1 (peroxisomal), Acyl-CoA dehydrogenase (medium-chain specific, mitochondrial), Acyl-CoA dehydrogenase (long-chain specific, mitochondrial), Acyl-coenzyme A oxidase 2 (peroxisomal), Acetyl-CoA acetyltransferase (mitochondrial), Acetyl-CoA acetyltransferase (cytosolic), Acyl-CoA dehydrogenase (short-chain specific, mitochondrial) and Trifunctional enzyme beta subunit (mitochondrial). |
|---|
| Structure | CCCC(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OCC1OC(C(O)C1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N InChI=1S/C25H42N7O17P3S/c1-4-5-16(34)53-9-8-27-15(33)6-7-28-23(37)20(36)25(2,3)11-46-52(43,44)49-51(41,42)45-10-14-19(48-50(38,39)40)18(35)24(47-14)32-13-31-17-21(26)29-12-30-22(17)32/h12-14,18-20,24,35-36H,4-11H2,1-3H3,(H,27,33)(H,28,37)(H,41,42)(H,43,44)(H2,26,29,30)(H2,38,39,40) |
|---|
| Synonyms | | Value | Source |
|---|
| Butanoyl-CoA | HMDB | | Butanoyl-coenzyme A | HMDB | | Butyryl-coenzyme A | HMDB |
|
|---|
| Chemical Formula | C25H42N7O17P3S |
|---|
| Average Molecular Weight | 837.624 |
|---|
| Monoisotopic Molecular Weight | 837.157073179 |
|---|
| IUPAC Name | {[5-(6-amino-9H-purin-9-yl)-2-[({[({3-[(2-{[2-(butanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]-3-hydroxy-2,2-dimethylpropoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-4-hydroxyoxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | butyryl-coa |
|---|
| CAS Registry Number | 2140-48-9 |
|---|
| SMILES | CCCC(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OCC1OC(C(O)C1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C25H42N7O17P3S/c1-4-5-16(34)53-9-8-27-15(33)6-7-28-23(37)20(36)25(2,3)11-46-52(43,44)49-51(41,42)45-10-14-19(48-50(38,39)40)18(35)24(47-14)32-13-31-17-21(26)29-12-30-22(17)32/h12-14,18-20,24,35-36H,4-11H2,1-3H3,(H,27,33)(H,28,37)(H,41,42)(H,43,44)(H2,26,29,30)(H2,38,39,40) |
|---|
| InChI Key | CRFNGMNYKDXRTN-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as acyl coas. These are organic compounds containing a coenzyme A substructure linked to an acyl chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | Acyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Pyrimidine
- Alkyl phosphate
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Azole
- Heteroaromatic compound
- Carbothioic s-ester
- Secondary alcohol
- Thiocarboxylic acid ester
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Organosulfur compound
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organic nitrogen compound
- Primary amine
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 3.52 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 10.2185 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 9.43 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 517.8 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatized |
|---|
| MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Butyryl-CoA 10V, Positive-QTOF | splash10-000i-1931000110-66d61bc46976c5d8c03b | 2016-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Butyryl-CoA 20V, Positive-QTOF | splash10-000j-1921000000-9cafa8d385e5954247c9 | 2016-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Butyryl-CoA 40V, Positive-QTOF | splash10-000j-1910000000-cd04baa0711dbfc497d6 | 2016-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Butyryl-CoA 10V, Negative-QTOF | splash10-017i-7920132450-45990a9ccc9ebd96868a | 2016-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Butyryl-CoA 20V, Negative-QTOF | splash10-001i-4910200010-a583bb4be14e1a89fbec | 2016-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Butyryl-CoA 40V, Negative-QTOF | splash10-056r-8900100000-88e7303e15d74ebe77cd | 2016-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Butyryl-CoA 10V, Negative-QTOF | splash10-000i-0000000090-073f428b330788172433 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Butyryl-CoA 20V, Negative-QTOF | splash10-014r-9200102230-8569a6d934e6ef361838 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Butyryl-CoA 40V, Negative-QTOF | splash10-0170-4003402900-aa858b6cf989ef7545e1 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Butyryl-CoA 10V, Positive-QTOF | splash10-000i-0000000190-722be0a11d66c5eb058c | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Butyryl-CoA 20V, Positive-QTOF | splash10-000i-0911001470-28ee98a4a04b37364813 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Butyryl-CoA 40V, Positive-QTOF | splash10-001i-0119000000-76181944bc6dba4c2ae5 | 2021-09-24 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum |
|
|---|
| General References | - Korman SH, Andresen BS, Zeharia A, Gutman A, Boneh A, Pitt JJ: 2-ethylhydracrylic aciduria in short/branched-chain acyl-CoA dehydrogenase deficiency: application to diagnosis and implications for the R-pathway of isoleucine oxidation. Clin Chem. 2005 Mar;51(3):610-7. Epub 2004 Dec 22. [PubMed:15615815 ]
- Gargus JJ, Boyle K, Bocian M, Roe DS, Vianey-Saban C, Roe CR: Respiratory complex II defect in siblings associated with a symptomatic secondary block in fatty acid oxidation. J Inherit Metab Dis. 2003;26(7):659-70. [PubMed:14707514 ]
- Fournet JC, Junien C: Genetics of congenital hyperinsulinism. Endocr Pathol. 2004 Fall;15(3):233-40. [PubMed:15640549 ]
- van Maldegem BT, Waterham HR, Duran M, van der Vlies M, van Woerden CS, Bobu LL, Wanders RJ, Wijburg FA: The 625G>A SCAD gene variant is common but not associated with increased C4-carnitine in newborn blood spots. J Inherit Metab Dis. 2005;28(4):557-62. [PubMed:15902559 ]
|
|---|