| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:49:09 UTC |
|---|
| HMDB ID | HMDB0001338 |
|---|
| Secondary Accession Numbers | - HMDB0059623
- HMDB01338
- HMDB59623
|
|---|
| Metabolite Identification |
|---|
| Common Name | Palmityl-CoA |
|---|
| Description | hexadecanoyl-CoA belongs to the class of organic compounds known as hexoses. These are monosaccharides in which the sugar unit is a is a six-carbon containing moeity. Thus, hexadecanoyl-CoA is considered to be a fatty ester lipid molecule. hexadecanoyl-CoA is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. Outside of the human body, hexadecanoyl-CoA has been detected, but not quantified in, several different foods, such as hedge mustards, swedes, nances, pak choy, and sacred lotus. This could make hexadecanoyl-CoA a potential biomarker for the consumption of these foods. |
|---|
| Structure | CCCCCCCCCCCCCCCC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N InChI=1S/C37H66N7O17P3S/c1-4-5-6-7-8-9-10-11-12-13-14-15-16-17-28(46)65-21-20-39-27(45)18-19-40-35(49)32(48)37(2,3)23-58-64(55,56)61-63(53,54)57-22-26-31(60-62(50,51)52)30(47)36(59-26)44-25-43-29-33(38)41-24-42-34(29)44/h24-26,30-32,36,47-48H,4-23H2,1-3H3,(H,39,45)(H,40,49)(H,53,54)(H,55,56)(H2,38,41,42)(H2,50,51,52)/t26-,30-,31-,32+,36-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| coenzyme A, S-Hexadecanoate | ChEBI | | Palmitoyl coenzyme A | ChEBI | | S-Palmitoylcoenzyme A | ChEBI | | coenzyme A, S-Hexadecanoic acid | Generator | | Hexadecanoyl-CoA | ChEBI | | CoA, Hexadecanoyl | MeSH | | CoA, Palmityl | MeSH | | Hexadecanoyl CoA | MeSH | | coenzyme A, Palmitoyl | MeSH | | Palmityl CoA | MeSH | | CoA, Palmitoyl | MeSH | | Palmitoyl CoA | MeSH | | Hexadecanoyl-coenzyme A | HMDB | | Palmityl coenzyme A | HMDB | | Palmityl-CoA | HMDB | | Palmityl-coenzyme A | HMDB | | S-Palmityl coenzyme A | HMDB | | S-Palmityl-CoA | HMDB | | S-Palmityl-coenzyme A | HMDB |
|
|---|
| Chemical Formula | C37H66N7O17P3S |
|---|
| Average Molecular Weight | 1005.943 |
|---|
| Monoisotopic Molecular Weight | 1005.344873947 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-2-({[({[(3R)-3-[(2-{[2-(hexadecanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]-3-hydroxy-2,2-dimethylpropoxy](hydroxy)phosphoryl}oxy)(hydroxy)phosphoryl]oxy}methyl)-4-hydroxyoxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | palmitoyl-coa |
|---|
| CAS Registry Number | 1763-10-6 |
|---|
| SMILES | CCCCCCCCCCCCCCCC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C37H66N7O17P3S/c1-4-5-6-7-8-9-10-11-12-13-14-15-16-17-28(46)65-21-20-39-27(45)18-19-40-35(49)32(48)37(2,3)23-58-64(55,56)61-63(53,54)57-22-26-31(60-62(50,51)52)30(47)36(59-26)44-25-43-29-33(38)41-24-42-34(29)44/h24-26,30-32,36,47-48H,4-23H2,1-3H3,(H,39,45)(H,40,49)(H,53,54)(H,55,56)(H2,38,41,42)(H2,50,51,52)/t26-,30-,31-,32+,36-/m1/s1 |
|---|
| InChI Key | MNBKLUUYKPBKDU-BBECNAHFSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as hexoses. These are monosaccharides in which the sugar unit is a is a six-carbon containing moeity. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Hexoses |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hexose monosaccharide
- Oxane
- Secondary alcohol
- Hemiacetal
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Hydrocarbon derivative
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 5.78 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 14.1244 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 4.61 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 245.5 seconds | 40023050 |
Predicted Kovats Retention IndicesNot Available |
|---|
| Spectra |
|---|
| MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Palmityl-CoA 10V, Positive-QTOF | splash10-000i-2709000101-7863d2df84c4c4e06bd2 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Palmityl-CoA 20V, Positive-QTOF | splash10-000i-1829200000-77fb81a8f15c5d2b616c | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Palmityl-CoA 40V, Positive-QTOF | splash10-000i-1914000000-016e1dd6b88c119f09b8 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Palmityl-CoA 10V, Negative-QTOF | splash10-0far-8971333504-cdf5697f5a74af8e63d1 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Palmityl-CoA 20V, Negative-QTOF | splash10-001r-2910301001-24e89da24748049880af | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Palmityl-CoA 40V, Negative-QTOF | splash10-056r-7900100000-2cd78f62ff0a7c2cb5b8 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Palmityl-CoA 10V, Negative-QTOF | splash10-0udi-9000000000-a5e30ac28e6aa4c5b77d | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Palmityl-CoA 20V, Negative-QTOF | splash10-0ug0-9010201301-1162e12d8cb42839e762 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Palmityl-CoA 40V, Negative-QTOF | splash10-004r-4102301209-063dede4760b705c02d9 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Palmityl-CoA 10V, Positive-QTOF | splash10-0a4i-9000000000-d041dea4fa9f52102bd4 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Palmityl-CoA 20V, Positive-QTOF | splash10-000i-3801000429-6b8dc383575e6c3d2b72 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Palmityl-CoA 40V, Positive-QTOF | splash10-0002-0001900000-a96e69193e5f6d651546 | 2021-09-24 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum |
|
|---|
| Biological Properties |
|---|
| Cellular Locations | |
|---|
| Biospecimen Locations | Not Available |
|---|
| Tissue Locations | - Brain
- Erythrocyte
- Fibroblasts
- Heart
- Kidney
- Liver
- Lung
- Neuron
- Pancreas
- Placenta
- Skeletal Muscle
|
|---|
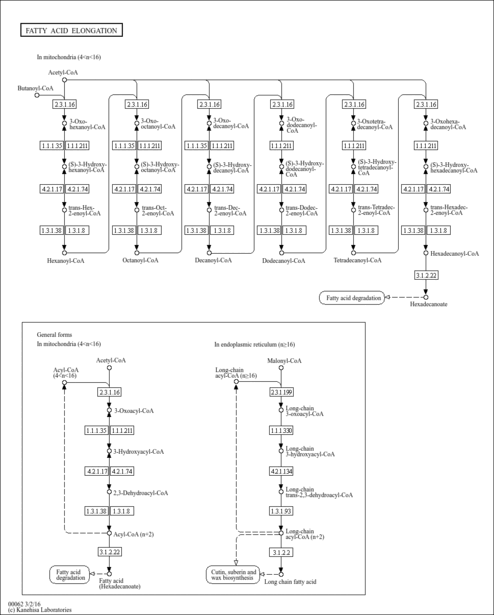
| Pathways | |
|---|
| Normal Concentrations |
|---|
| Not Available |
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | None |
|---|
| Associated OMIM IDs | None |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB022562 |
|---|
| KNApSAcK ID | C00007462 |
|---|
| Chemspider ID | 559149 |
|---|
| KEGG Compound ID | C00154 |
|---|
| BioCyc ID | PALMITYL-COA |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Not Available |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 644109 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15525 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | Not Available |
|---|
| MarkerDB ID | Not Available |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Not Available |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| General References | - Bajaj M, Suraamornkul S, Romanelli A, Cline GW, Mandarino LJ, Shulman GI, DeFronzo RA: Effect of a sustained reduction in plasma free fatty acid concentration on intramuscular long-chain fatty Acyl-CoAs and insulin action in type 2 diabetic patients. Diabetes. 2005 Nov;54(11):3148-53. [PubMed:16249438 ]
- Gohil K, Jones DA, Edwards RH: Fatty acid oxidation in mitochondria from needle biopsy samples of human skeletal muscle. Clin Sci (Lond). 1984 Feb;66(2):173-8. [PubMed:6319070 ]
- Berge RK, Hagen LE, Farstad M: Isolation of palmitoyl-CoA hydrolases from human blood platelets. Biochem J. 1981 Dec 1;199(3):639-47. [PubMed:6122441 ]
- Casteels M, Schepers L, Parmentier G, Eyssen HJ, Mannaerts GP: Activation and peroxisomal beta-oxidation of fatty acids and bile acid intermediates in liver from Bombina orientalis and from the rat. Comp Biochem Physiol B. 1989;92(1):129-32. [PubMed:2706931 ]
- Carroll JE, McGuire BS, Hall CL: Fatty acyl-CoA dehydrogenase enzymes in human skeletal muscle. Clin Chim Acta. 1986 Dec 30;161(3):327-33. [PubMed:3802539 ]
- Holloway GP, Bezaire V, Heigenhauser GJ, Tandon NN, Glatz JF, Luiken JJ, Bonen A, Spriet LL: Mitochondrial long chain fatty acid oxidation, fatty acid translocase/CD36 content and carnitine palmitoyltransferase I activity in human skeletal muscle during aerobic exercise. J Physiol. 2006 Feb 15;571(Pt 1):201-10. Epub 2005 Dec 15. [PubMed:16357012 ]
- Bakken AM, Farstad M: Identical subcellular distribution of palmitoyl-CoA and arachidonoyl-CoA synthetase activities in human blood platelets. Biochem J. 1989 Jul 1;261(1):71-6. [PubMed:2528345 ]
- Fukao T, Watanabe H, Orii K, Takahashi Y, Hirano A, Kondo T, Yamaguchi S, Aoyama T, Kondo N: Myopathic form of very-long chain acyl-coa dehydrogenase deficiency: evidence for temperature-sensitive mild mutations in both mutant alleles in a Japanese girl. Pediatr Res. 2001 Feb;49(2):227-31. [PubMed:11158518 ]
- Haughey NJ, Cutler RG, Tamara A, McArthur JC, Vargas DL, Pardo CA, Turchan J, Nath A, Mattson MP: Perturbation of sphingolipid metabolism and ceramide production in HIV-dementia. Ann Neurol. 2004 Feb;55(2):257-67. [PubMed:14755730 ]
- Tonsgard JH, Stephens JK, Rhead WJ, Penn D, Horwitz AL, Kirschner BS, Whitington PF, Berger S, Tripp ME: Defect in fatty acid oxidation: laboratory and pathologic findings in a patient. Pediatr Neurol. 1991 Mar-Apr;7(2):125-30. [PubMed:2059253 ]
- Wanders RJ, van Roermund CW, de Vries CT, van den Bosch H, Schrakamp G, Tager JM, Schram AW, Schutgens RB: Peroxisomal beta-oxidation of palmitoyl-CoA in human liver homogenates and its deficiency in the cerebro-hepato-renal (Zellweger) syndrome. Clin Chim Acta. 1986 Aug 30;159(1):1-10. [PubMed:2944672 ]
- Simons K, Toomre D: Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000 Oct;1(1):31-9. [PubMed:11413487 ]
- Watson AD: Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J Lipid Res. 2006 Oct;47(10):2101-11. Epub 2006 Aug 10. [PubMed:16902246 ]
- Sethi JK, Vidal-Puig AJ: Thematic review series: adipocyte biology. Adipose tissue function and plasticity orchestrate nutritional adaptation. J Lipid Res. 2007 Jun;48(6):1253-62. Epub 2007 Mar 20. [PubMed:17374880 ]
- Lingwood D, Simons K: Lipid rafts as a membrane-organizing principle. Science. 2010 Jan 1;327(5961):46-50. doi: 10.1126/science.1174621. [PubMed:20044567 ]
- Gunstone, Frank D., John L. Harwood, and Albert J. Dijkstra (2007). The lipid handbook with CD-ROM. CRC Press.
|
|---|