| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2006-02-16 11:10:57 UTC |
|---|
| Update Date | 2022-03-07 02:49:10 UTC |
|---|
| HMDB ID | HMDB0001852 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | all-trans-Retinoic acid |
|---|
| Description | all-trans-Retinoic acid is an isomer of retinoic acid, the oxidized form of vitamin A. Retinoic acid functions in determining position along embryonic anterior/posterior axis in chordates. It acts through Hox genes, which ultimately controls anterior/posterior patterning in early developmental stages (PMID:17495912 ). It is an important regulator of gene expression during growth and development, and in neoplasms. As a drug, all-trans-retinoic acid is known as tretinoin. Tretinoin is derived from maternal vitamin A and is essential for normal growth and embryonic development. An excess of tretinoin can be teratogenic. Tretinoin is used in the treatment of psoriasis, acne vulgaris, and several other skin diseases. It has also been approved for use in promyelocytic leukemia (leukemia, promyelocytic, acute). |
|---|
| Structure | C\C(\C=C\C1=C(C)CCCC1(C)C)=C/C=C/C(/C)=C/C(O)=O InChI=1S/C20H28O2/c1-15(8-6-9-16(2)14-19(21)22)11-12-18-17(3)10-7-13-20(18,4)5/h6,8-9,11-12,14H,7,10,13H2,1-5H3,(H,21,22)/b9-6+,12-11+,15-8+,16-14+ |
|---|
| Synonyms | | Value | Source |
|---|
| (all-e)-3,7-Dimethyl-9-(2,6,6-trimethyl-1-cyclohexen-1-yl)-2,4,6,8-nonatetraenoic acid | ChEBI | | 3,7-Dimethyl-9-(2,6,6-trimethyl-1-cyclohexene-1-yl)-2,4,6,8-nonatetraenoic acid (ecl) | ChEBI | | AGN 100335 | ChEBI | | all-(e)-Retinoic acid | ChEBI | | all-trans-beta-Retinoic acid | ChEBI | | all-trans-Tretinoin | ChEBI | | all-trans-Vitamin a acid | ChEBI | | all-trans-Vitamin a1 acid | ChEBI | | beta-Retinoic acid | ChEBI | | Eudyna | ChEBI | | Renova | ChEBI | | Retin-a | ChEBI | | Retinoic acid | ChEBI | | Retisol-a | ChEBI | | Ro 1-5488 | ChEBI | | Solage | ChEBI | | Stieva-a | ChEBI | | trans-Retinoic acid | ChEBI | | Tretin m | ChEBI | | Tretinoin | ChEBI | | Vesanoid | ChEBI | | Vitamin a acid | ChEBI | | Vitinoin | ChEBI | | all-trans-Retinoate | Kegg | | Avita | Kegg | | Retin a | Kegg | | (all-e)-3,7-Dimethyl-9-(2,6,6-trimethyl-1-cyclohexen-1-yl)-2,4,6,8-nonatetraenoate | Generator | | 3,7-Dimethyl-9-(2,6,6-trimethyl-1-cyclohexene-1-yl)-2,4,6,8-nonatetraenoate (ecl) | Generator | | all-(e)-Retinoate | Generator | | all-trans-b-Retinoate | Generator | | all-trans-b-Retinoic acid | Generator | | all-trans-beta-Retinoate | Generator | | all-trans-Β-retinoate | Generator | | all-trans-Β-retinoic acid | Generator | | b-Retinoate | Generator | | b-Retinoic acid | Generator | | beta-Retinoate | Generator | | Β-retinoate | Generator | | Β-retinoic acid | Generator | | Retinoate | Generator | | trans-Retinoate | Generator | | (2E,4E,6E,8E)-3,7-Dimethyl-9-(2,6,6-trimethylcyclohexen-1-yl)nona-2,4,6,8-tetraenoate | HMDB | | Tretinoin zinc salt | HMDB | | trans Retinoic acid | HMDB | | Acid, all-trans-retinoic | HMDB | | Salt, tretinoin sodium | HMDB | | beta all trans Retinoic acid | HMDB | | beta-all-trans-Retinoic acid | HMDB | | Acid, vitamin a | HMDB | | Potassium salt, tretinoin | HMDB | | Sodium salt, tretinoin | HMDB | | Acid, beta-all-trans-retinoic | HMDB | | Tretinoin potassium salt | HMDB | | Zinc salt, tretinoin | HMDB | | Acid, retinoic | HMDB | | Acid, trans-retinoic | HMDB | | Salt, tretinoin potassium | HMDB | | Salt, tretinoin zinc | HMDB | | Tretinoin sodium salt | HMDB | | all trans Retinoic acid | HMDB | | Retin-a micro | HMDB | | Atralin | HMDB | | Tretin-X | HMDB | | all-trans-Retinoic acid | MeSH |
|
|---|
| Chemical Formula | C20H28O2 |
|---|
| Average Molecular Weight | 300.442 |
|---|
| Monoisotopic Molecular Weight | 300.208930142 |
|---|
| IUPAC Name | (2E,4E,6E,8E)-3,7-dimethyl-9-(2,6,6-trimethylcyclohex-1-en-1-yl)nona-2,4,6,8-tetraenoic acid |
|---|
| Traditional Name | tretinoin |
|---|
| CAS Registry Number | 302-79-4 |
|---|
| SMILES | C\C(\C=C\C1=C(C)CCCC1(C)C)=C/C=C/C(/C)=C/C(O)=O |
|---|
| InChI Identifier | InChI=1S/C20H28O2/c1-15(8-6-9-16(2)14-19(21)22)11-12-18-17(3)10-7-13-20(18,4)5/h6,8-9,11-12,14H,7,10,13H2,1-5H3,(H,21,22)/b9-6+,12-11+,15-8+,16-14+ |
|---|
| InChI Key | SHGAZHPCJJPHSC-YCNIQYBTSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as retinoids. These are oxygenated derivatives of 3,7-dimethyl-1-(2,6,6-trimethylcyclohex-1-enyl)nona-1,3,5,7-tetraene and derivatives thereof. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Retinoids |
|---|
| Direct Parent | Retinoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Retinoic acid
- Diterpenoid
- Retinoid skeleton
- Medium-chain fatty acid
- Branched fatty acid
- Methyl-branched fatty acid
- Fatty acyl
- Unsaturated fatty acid
- Fatty acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Hydrocarbon derivative
- Organooxygen compound
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 181 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | 6.30 | HANSCH,C ET AL. (1995) |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter)). Predicted by Afia on May 17, 2022. | 8.33 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 25.9972 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 1.19 minutes | 32390414 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 3405.6 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 798.4 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 300.1 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 502.7 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 379.8 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 1030.3 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 960.1 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 99.1 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 2162.1 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 747.4 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 1810.2 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 825.2 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 576.6 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 507.5 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 760.6 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 8.5 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatizedDerivatized |
|---|
| Spectra |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - all-trans-Retinoic acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-000i-2090000000-db078f48efb8038384f2 | 2018-04-09 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - all-trans-Retinoic acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid Quattro_QQQ 10V, Positive-QTOF (Annotated) | splash10-0udi-0009000000-12dbd83959268dee6dbf | 2018-05-28 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid Quattro_QQQ 25V, Positive-QTOF (Annotated) | splash10-066r-4900000000-31c9262cbbf0bad5b738 | 2018-05-28 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid Quattro_QQQ 40V, Positive-QTOF (Annotated) | splash10-0fsl-9600000000-c6681587f0a9ae7a712e | 2018-05-28 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid 20V, Positive-QTOF | splash10-0mc0-4941000000-401db468e6db7c5bbb5a | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid 35V, Negative-QTOF | splash10-0a4i-0090000000-d05c305f987a713dfaa0 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid 10V, Positive-QTOF | splash10-0udi-0239000000-09da873ecbe1e09a1602 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid 10V, Negative-QTOF | splash10-0002-0090000000-dca272cadb8a3ba3f819 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid 40V, Negative-QTOF | splash10-016r-9630000000-553ddaf9452713df67ee | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid 20V, Negative-QTOF | splash10-0a4i-0390000000-241af7f979164ecfbf34 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid 20V, Positive-QTOF | splash10-0mc0-4941000000-05bed4117d9ed953a2ca | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid 40V, Positive-QTOF | splash10-001i-9400000000-3ecfeb7f6c8384d2a0d3 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid 10V, Positive-QTOF | splash10-0udi-0239000000-2c6852256fcbebe1a2aa | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - all-trans-Retinoic acid 35V, Positive-QTOF | splash10-00di-0910000000-a1c1aa8b35e78558cd5b | 2021-09-20 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Retinoic acid 10V, Positive-QTOF | splash10-0uei-0494000000-28e6366fdd47e8ba2117 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Retinoic acid 20V, Positive-QTOF | splash10-052r-2980000000-3e7fc78de83c23830416 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Retinoic acid 40V, Positive-QTOF | splash10-052r-5900000000-ef4d86f8bcf86959f794 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Retinoic acid 10V, Negative-QTOF | splash10-052b-0090000000-e07ffb4c1e5c63ab1b59 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Retinoic acid 20V, Negative-QTOF | splash10-052b-0090000000-a0a8411213bbd2543243 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Retinoic acid 40V, Negative-QTOF | splash10-000i-3690000000-97e6a74798dd69c30ad1 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Retinoic acid 10V, Positive-QTOF | splash10-0zir-0973000000-3040e9e4a1a5448efbcb | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Retinoic acid 20V, Positive-QTOF | splash10-060c-1950000000-f3b9c9b378f60b8c49a9 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Retinoic acid 40V, Positive-QTOF | splash10-0ar3-5910000000-de22eb65f274483f3f98 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Retinoic acid 10V, Negative-QTOF | splash10-052b-0090000000-5f942e3b870e7bd0121b | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Retinoic acid 20V, Negative-QTOF | splash10-0a4i-0190000000-22b445382d3977280045 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Retinoic acid 40V, Negative-QTOF | splash10-016r-4900000000-0005b37f6d54a42a84f6 | 2021-09-24 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CDCl3, experimental) | 2018-05-28 | Wishart Lab | View Spectrum |
|
|---|
| Biological Properties |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Membrane (predicted from logP)
- Nucleus
- Endoplasmic reticulum
|
|---|
| Biospecimen Locations | |
|---|
| Tissue Locations | - Adipose Tissue
- Adrenal Cortex
- Adrenal Gland
- Bladder
- Brain
- Epidermis
- Fibroblasts
- Intestine
- Kidney
- Liver
- Lung
- Neuron
- Pancreas
- Placenta
- Platelet
- Prostate
- Spleen
- Testis
|
|---|
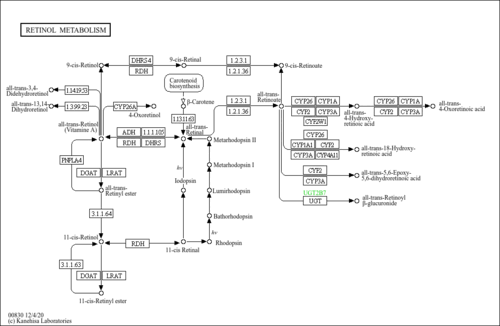
| Pathways | |
|---|
| Normal Concentrations |
|---|
| |
| Blood | Detected and Quantified | 0.20 +/- 0.18 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 0.0145 +/- 0.0092 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details |
|
|---|
| Abnormal Concentrations |
|---|
| Not Available |
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | None |
|---|
| Associated OMIM IDs | None |
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB022710 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 392618 |
|---|
| KEGG Compound ID | C00777 |
|---|
| BioCyc ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Tretinoin |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 444795 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15367 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | RETN |
|---|
| MarkerDB ID | Not Available |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Solladie, Guy; Girardin, Andre. Highly stereoselective synthesis of vitamin A and all-trans retinoic acid by low-valent titanium induced reductive elimination. Tetrahedron Letters (1988), 29(2), 213-16. |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| General References | - Czernik PJ, Little JM, Barone GW, Raufman JP, Radominska-Pandya A: Glucuronidation of estrogens and retinoic acid and expression of UDP-glucuronosyltransferase 2B7 in human intestinal mucosa. Drug Metab Dispos. 2000 Oct;28(10):1210-6. [PubMed:10997942 ]
- Barua AB, Olson JA: Retinoyl beta-glucuronide: an endogenous compound of human blood. Am J Clin Nutr. 1986 Apr;43(4):481-5. [PubMed:3962900 ]
- Napoli JL, Posch KP, Fiorella PD, Boerman MH: Physiological occurrence, biosynthesis and metabolism of retinoic acid: evidence for roles of cellular retinol-binding protein (CRBP) and cellular retinoic acid-binding protein (CRABP) in the pathway of retinoic acid homeostasis. Biomed Pharmacother. 1991;45(4-5):131-43. [PubMed:1932598 ]
- Giannini G, Di Marcotullio L, Ristori E, Zani M, Crescenzi M, Scarpa S, Piaggio G, Vacca A, Peverali FA, Diana F, Screpanti I, Frati L, Gulino A: HMGI(Y) and HMGI-C genes are expressed in neuroblastoma cell lines and tumors and affect retinoic acid responsiveness. Cancer Res. 1999 May 15;59(10):2484-92. [PubMed:10344762 ]
- Lee YF, Bao BY, Chang C: Modulation of the retinoic acid-induced cell apoptosis and differentiation by the human TR4 orphan nuclear receptor. Biochem Biophys Res Commun. 2004 Oct 22;323(3):876-83. [PubMed:15381082 ]
- Mercader J, Ribot J, Murano I, Felipe F, Cinti S, Bonet ML, Palou A: Remodeling of white adipose tissue after retinoic acid administration in mice. Endocrinology. 2006 Nov;147(11):5325-32. Epub 2006 Jul 13. [PubMed:16840543 ]
- Natsume N, Kondo S, Matsuyama Y, Sumida K, Inou H, Kawakami N, Sandell LJ, Iwata H: Analysis of cartilage-derived retinoic acid-sensitive protein in cerebrospinal fluid From patients With spinal diseases. Spine (Phila Pa 1976). 2001 Jan 15;26(2):157-60. [PubMed:11154535 ]
- Baron JM, Heise R, Blaner WS, Neis M, Joussen S, Dreuw A, Marquardt Y, Saurat JH, Merk HF, Bickers DR, Jugert FK: Retinoic acid and its 4-oxo metabolites are functionally active in human skin cells in vitro. J Invest Dermatol. 2005 Jul;125(1):143-53. [PubMed:15982314 ]
- Paez-Pereda M, Kovalovsky D, Hopfner U, Theodoropoulou M, Pagotto U, Uhl E, Losa M, Stalla J, Grubler Y, Missale C, Arzt E, Stalla GK: Retinoic acid prevents experimental Cushing syndrome. J Clin Invest. 2001 Oct;108(8):1123-31. [PubMed:11602619 ]
- Saito S, Kondo S, Mishima S, Ishiguro N, Hasegawa Y, Sandell LJ, Iwata H: Analysis of cartilage-derived retinoic-acid-sensitive protein (CD-RAP) in synovial fluid from patients with osteoarthritis and rheumatoid arthritis. J Bone Joint Surg Br. 2002 Sep;84(7):1066-9. [PubMed:12358374 ]
- Meyer E, Lambert WE, De Leenheer AP: Simultaneous determination of endogenous retinoic acid isomers and retinol in human plasma by isocratic normal-phase HPLC with ultraviolet detection. Clin Chem. 1994 Jan;40(1):48-57. [PubMed:8287543 ]
- Masgrau-Peya E, Salomon D, Saurat JH, Meda P: In vivo modulation of connexins 43 and 26 of human epidermis by topical retinoic acid treatment. J Histochem Cytochem. 1997 Sep;45(9):1207-15. [PubMed:9283608 ]
- Lopez-Boado YS, Klaus M, Dawson MI, Lopez-Otin C: Retinoic acid-induced expression of apolipoprotein D and concomitant growth arrest in human breast cancer cells are mediated through a retinoic acid receptor RARalpha-dependent signaling pathway. J Biol Chem. 1996 Dec 13;271(50):32105-11. [PubMed:8943263 ]
- Stewart ME, Benoit AM, Downing DT, Strauss JS: Suppression of sebum secretion with 13-cis-retinoic acid: effect on individual skin surface lipids and implications for their anatomic origin. J Invest Dermatol. 1984 Jan;82(1):74-8. [PubMed:6228612 ]
- Tse HK, Leung MB, Woolf AS, Menke AL, Hastie ND, Gosling JA, Pang CP, Shum AS: Implication of Wt1 in the pathogenesis of nephrogenic failure in a mouse model of retinoic acid-induced caudal regression syndrome. Am J Pathol. 2005 May;166(5):1295-307. [PubMed:15855632 ]
- Patel JB, Huynh CK, Handratta VD, Gediya LK, Brodie AM, Goloubeva OG, Clement OO, Nanne IP, Soprano DR, Njar VC: Novel retinoic acid metabolism blocking agents endowed with multiple biological activities are efficient growth inhibitors of human breast and prostate cancer cells in vitro and a human breast tumor xenograft in nude mice. J Med Chem. 2004 Dec 30;47(27):6716-29. [PubMed:15615521 ]
- Thiboutot D, Martin P, Volikos L, Gilliland K: Oxidative activity of the type 2 isozyme of 17beta-hydroxysteroid dehydrogenase (17beta-HSD) predominates in human sebaceous glands. J Invest Dermatol. 1998 Sep;111(3):390-5. [PubMed:9740229 ]
- Holland LZ: Developmental biology: a chordate with a difference. Nature. 2007 May 10;447(7141):153-5. [PubMed:17495912 ]
|
|---|