| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2006-05-22 15:12:14 UTC |
|---|
| Update Date | 2022-03-07 02:49:17 UTC |
|---|
| HMDB ID | HMDB0002845 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Hexanoyl-CoA |
|---|
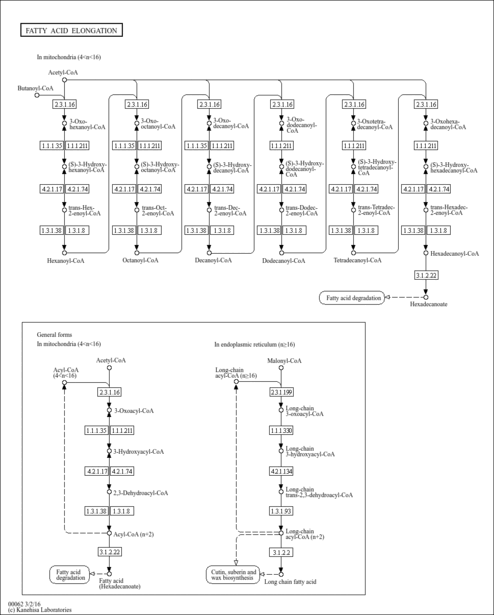
| Description | Hexanoyl-CoA, also known as hexanoyl-coenzyme A or caproyl-CoA, is a medium-chain fatty acyl-CoA having hexanoyl as the acyl group. Hexanoyl-CoA is slightly soluble (in water) and an extremely strong acidic compound (based on its pKa). Within the cell, hexanoyl-CoA is primarily located in the membrane (predicted from logP). It can also be found in the extracellular space. Hexanoyl-CoA exists in all living organisms, ranging from bacteria to humans. In humans, hexanoyl-CoA is involved in the biosynthesis and oxidation of fatty acids as well as in ceramide formation. Hexanoyl-CoA is also involved in few metabolic disorders, such as fatty acid elongation in mitochondria, mitochondrial beta-oxidation of medium chain saturated fatty acids, and mitochondrial beta-oxidation of short chain saturated fatty acids. |
|---|
| Structure | CCCCCC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N InChI=1S/C27H46N7O17P3S/c1-4-5-6-7-18(36)55-11-10-29-17(35)8-9-30-25(39)22(38)27(2,3)13-48-54(45,46)51-53(43,44)47-12-16-21(50-52(40,41)42)20(37)26(49-16)34-15-33-19-23(28)31-14-32-24(19)34/h14-16,20-22,26,37-38H,4-13H2,1-3H3,(H,29,35)(H,30,39)(H,43,44)(H,45,46)(H2,28,31,32)(H2,40,41,42)/t16-,20-,21-,22+,26-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| Caproyl-CoA | ChEBI | | Caproyl-coenzyme A | ChEBI | | coenzyme A, S-Hexanoate | ChEBI | | Hexanoyl-coenzyme A | ChEBI | | N-Hexanoyl-CoA | ChEBI | | N-Hexanoyl-coenzyme A | ChEBI | | S-Hexanoyl-CoA | ChEBI | | S-Hexanoyl-coenzym-a | ChEBI | | S-Hexanoyl-coenzyme A | ChEBI | | coenzyme A, S-Hexanoic acid | Generator | | CoA(6:0) | HMDB |
|
|---|
| Chemical Formula | C27H46N7O17P3S |
|---|
| Average Molecular Weight | 865.677 |
|---|
| Monoisotopic Molecular Weight | 865.188373307 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-2-({[({[(3R)-3-[(2-{[2-(hexanoylsulfanyl)ethyl]carbamoyl}ethyl)carbamoyl]-3-hydroxy-2,2-dimethylpropoxy](hydroxy)phosphoryl}oxy)(hydroxy)phosphoryl]oxy}methyl)-4-hydroxyoxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | hexanoyl-coa |
|---|
| CAS Registry Number | 5060-32-2 |
|---|
| SMILES | CCCCCC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C27H46N7O17P3S/c1-4-5-6-7-18(36)55-11-10-29-17(35)8-9-30-25(39)22(38)27(2,3)13-48-54(45,46)51-53(43,44)47-12-16-21(50-52(40,41)42)20(37)26(49-16)34-15-33-19-23(28)31-14-32-24(19)34/h14-16,20-22,26,37-38H,4-13H2,1-3H3,(H,29,35)(H,30,39)(H,43,44)(H,45,46)(H2,28,31,32)(H2,40,41,42)/t16-,20-,21-,22+,26-/m1/s1 |
|---|
| InChI Key | OEXFMSFODMQEPE-HDRQGHTBSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as 2,3,4-saturated fatty acyl coas. These are acyl-CoAs carrying a 2,3,4-saturated fatty acyl chain. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | 2,3,4-saturated fatty acyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Pyrimidine
- Alkyl phosphate
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Azole
- Heteroaromatic compound
- Carbothioic s-ester
- Secondary alcohol
- Thiocarboxylic acid ester
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Organosulfur compound
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organic nitrogen compound
- Primary amine
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatized |
|---|
| MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hexanoyl-CoA 10V, Positive-QTOF | splash10-000j-1931000110-940fe882d4b8cefb1914 | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hexanoyl-CoA 20V, Positive-QTOF | splash10-0079-1921000000-de60fcd8bc6f0c9bf08d | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hexanoyl-CoA 40V, Positive-QTOF | splash10-0079-1910000000-29936f42a08315dcc8ff | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hexanoyl-CoA 10V, Negative-QTOF | splash10-01rt-7911042450-6db5d278813c82f99785 | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hexanoyl-CoA 20V, Negative-QTOF | splash10-001i-4901110010-64b9e09e0fd795b938c8 | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hexanoyl-CoA 40V, Negative-QTOF | splash10-056r-8900100000-f1097e6a0dd8990fbfb7 | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hexanoyl-CoA 10V, Negative-QTOF | splash10-03di-0000000090-5cc3f6bd431e230faf4a | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hexanoyl-CoA 20V, Negative-QTOF | splash10-03gj-9600204660-0d998b9d3d0254c489f4 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hexanoyl-CoA 40V, Negative-QTOF | splash10-00p0-7104802930-08e206f3a7fa95fa94d3 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hexanoyl-CoA 10V, Positive-QTOF | splash10-014i-0000000190-fde2b5c220974cde750c | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hexanoyl-CoA 20V, Positive-QTOF | splash10-000i-1901001680-96f2a80cee26fb36881b | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Hexanoyl-CoA 40V, Positive-QTOF | splash10-0a4i-0119000000-74dda344be4f5c2e3b01 | 2021-09-22 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum |
|
|---|