| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2006-05-22 15:12:32 UTC |
|---|
| Update Date | 2023-02-21 17:16:32 UTC |
|---|
| HMDB ID | HMDB0003099 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 1-Methyluric acid |
|---|
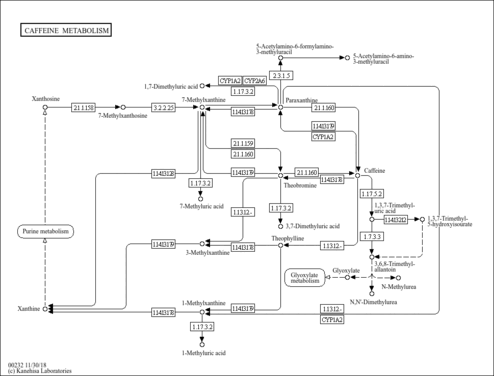
| Description | 1-Methyluric acid is one of the three main theophylline metabolites in man. 1-Methyluric acid is one of the purine components in urinary calculi. Methylated purines originate from the metabolism of methylxanthines (caffeine, theophylline, and theobromine). Methyluric acids can be distinguished from uric acid via simple methods routinely used in clinical laboratories, requiring the use of high-performance liquid chromatography (HPLC). Purine derivatives in urinary calculi could be considered markers of abnormal purine metabolism. The content of a purine derivative in stone depends on its average urinary excretion in the general population, similarity to the chemical structure of uric acid, and content of the latter in stone. This suggests that purines in stones represent a solid solution with uric acid as solvent. It is also plausible that methylxanthines, ubiquitous components of the diet and drugs, are involved in the pathogenesis of urolithiasis. Caffeine is metabolized via successive pathways mainly catalyzed by CYP1A2, xanthine oxidase, or N-acetyltransferase-2 to give 14 different metabolites. CYP1A2 activity shows an inter-individual variability among the population. CYP1A2, an isoform of the CYP1A cytochrome P450 superfamily, is involved in the metabolism of many drugs and plays a potentially important role in the induction of chemical carcinogenesis (PMID:11712316 , 15833286 , 3506820 , 15013152 , 4039734 , 9890610 ). |
|---|
| Structure | CN1C(=O)NC2=C(NC(=O)N2)C1=O InChI=1S/C6H6N4O3/c1-10-4(11)2-3(9-6(10)13)8-5(12)7-2/h1H3,(H,9,13)(H2,7,8,12) |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Methylate | Generator | | 1-Methylic acid | Generator | | 1-Methylurate | HMDB, MeSH |
|
|---|
| Chemical Formula | C6H6N4O3 |
|---|
| Average Molecular Weight | 182.1368 |
|---|
| Monoisotopic Molecular Weight | 182.043990078 |
|---|
| IUPAC Name | 1-methyl-2,3,6,7,8,9-hexahydro-1H-purine-2,6,8-trione |
|---|
| Traditional Name | 1-methyluric acid |
|---|
| CAS Registry Number | 708-79-2 |
|---|
| SMILES | CN1C(=O)NC2=C(NC(=O)N2)C1=O |
|---|
| InChI Identifier | InChI=1S/C6H6N4O3/c1-10-4(11)2-3(9-6(10)13)8-5(12)7-2/h1H3,(H,9,13)(H2,7,8,12) |
|---|
| InChI Key | QFDRTQONISXGJA-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Xanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthine
- 6-oxopurine
- Purinone
- Alkaloid or derivatives
- Pyrimidone
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Vinylogous amide
- Lactam
- Urea
- Azacycle
- Hydrocarbon derivative
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 5 mg/mL | Not Available | | LogP | -0.57 | GASPARI,F & BONATI,M (1987) |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 1.97 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 9.2463 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 2.71 minutes | 32390414 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 444.4 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 323.2 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 61.7 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 197.0 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 39.3 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 277.3 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 250.8 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 614.1 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 596.3 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 67.3 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 676.2 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 186.9 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 246.2 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 705.7 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 366.9 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 285.2 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| 1-Methyluric acid,1TMS,isomer #1 | CN1C(=O)C2=C([NH]C(=O)[NH]2)N([Si](C)(C)C)C1=O | 2011.6 | Semi standard non polar | 33892256 | | 1-Methyluric acid,1TMS,isomer #1 | CN1C(=O)C2=C([NH]C(=O)[NH]2)N([Si](C)(C)C)C1=O | 2235.8 | Standard non polar | 33892256 | | 1-Methyluric acid,1TMS,isomer #1 | CN1C(=O)C2=C([NH]C(=O)[NH]2)N([Si](C)(C)C)C1=O | 3042.6 | Standard polar | 33892256 | | 1-Methyluric acid,1TMS,isomer #2 | CN1C(=O)[NH]C2=C(C1=O)N([Si](C)(C)C)C(=O)[NH]2 | 2036.5 | Semi standard non polar | 33892256 | | 1-Methyluric acid,1TMS,isomer #2 | CN1C(=O)[NH]C2=C(C1=O)N([Si](C)(C)C)C(=O)[NH]2 | 2221.8 | Standard non polar | 33892256 | | 1-Methyluric acid,1TMS,isomer #2 | CN1C(=O)[NH]C2=C(C1=O)N([Si](C)(C)C)C(=O)[NH]2 | 2945.6 | Standard polar | 33892256 | | 1-Methyluric acid,1TMS,isomer #3 | CN1C(=O)[NH]C2=C([NH]C(=O)N2[Si](C)(C)C)C1=O | 2008.5 | Semi standard non polar | 33892256 | | 1-Methyluric acid,1TMS,isomer #3 | CN1C(=O)[NH]C2=C([NH]C(=O)N2[Si](C)(C)C)C1=O | 2268.2 | Standard non polar | 33892256 | | 1-Methyluric acid,1TMS,isomer #3 | CN1C(=O)[NH]C2=C([NH]C(=O)N2[Si](C)(C)C)C1=O | 3068.8 | Standard polar | 33892256 | | 1-Methyluric acid,2TMS,isomer #1 | CN1C(=O)C2=C([NH]C(=O)N2[Si](C)(C)C)N([Si](C)(C)C)C1=O | 2016.0 | Semi standard non polar | 33892256 | | 1-Methyluric acid,2TMS,isomer #1 | CN1C(=O)C2=C([NH]C(=O)N2[Si](C)(C)C)N([Si](C)(C)C)C1=O | 2237.6 | Standard non polar | 33892256 | | 1-Methyluric acid,2TMS,isomer #1 | CN1C(=O)C2=C([NH]C(=O)N2[Si](C)(C)C)N([Si](C)(C)C)C1=O | 2509.3 | Standard polar | 33892256 | | 1-Methyluric acid,2TMS,isomer #2 | CN1C(=O)C2=C(N([Si](C)(C)C)C(=O)[NH]2)N([Si](C)(C)C)C1=O | 1990.6 | Semi standard non polar | 33892256 | | 1-Methyluric acid,2TMS,isomer #2 | CN1C(=O)C2=C(N([Si](C)(C)C)C(=O)[NH]2)N([Si](C)(C)C)C1=O | 2296.2 | Standard non polar | 33892256 | | 1-Methyluric acid,2TMS,isomer #2 | CN1C(=O)C2=C(N([Si](C)(C)C)C(=O)[NH]2)N([Si](C)(C)C)C1=O | 2602.1 | Standard polar | 33892256 | | 1-Methyluric acid,2TMS,isomer #3 | CN1C(=O)[NH]C2=C(C1=O)N([Si](C)(C)C)C(=O)N2[Si](C)(C)C | 2001.2 | Semi standard non polar | 33892256 | | 1-Methyluric acid,2TMS,isomer #3 | CN1C(=O)[NH]C2=C(C1=O)N([Si](C)(C)C)C(=O)N2[Si](C)(C)C | 2268.7 | Standard non polar | 33892256 | | 1-Methyluric acid,2TMS,isomer #3 | CN1C(=O)[NH]C2=C(C1=O)N([Si](C)(C)C)C(=O)N2[Si](C)(C)C | 2562.0 | Standard polar | 33892256 | | 1-Methyluric acid,3TMS,isomer #1 | CN1C(=O)C2=C(N([Si](C)(C)C)C1=O)N([Si](C)(C)C)C(=O)N2[Si](C)(C)C | 2126.4 | Semi standard non polar | 33892256 | | 1-Methyluric acid,3TMS,isomer #1 | CN1C(=O)C2=C(N([Si](C)(C)C)C1=O)N([Si](C)(C)C)C(=O)N2[Si](C)(C)C | 2280.0 | Standard non polar | 33892256 | | 1-Methyluric acid,3TMS,isomer #1 | CN1C(=O)C2=C(N([Si](C)(C)C)C1=O)N([Si](C)(C)C)C(=O)N2[Si](C)(C)C | 2287.9 | Standard polar | 33892256 | | 1-Methyluric acid,1TBDMS,isomer #1 | CN1C(=O)C2=C([NH]C(=O)[NH]2)N([Si](C)(C)C(C)(C)C)C1=O | 2198.5 | Semi standard non polar | 33892256 | | 1-Methyluric acid,1TBDMS,isomer #1 | CN1C(=O)C2=C([NH]C(=O)[NH]2)N([Si](C)(C)C(C)(C)C)C1=O | 2464.4 | Standard non polar | 33892256 | | 1-Methyluric acid,1TBDMS,isomer #1 | CN1C(=O)C2=C([NH]C(=O)[NH]2)N([Si](C)(C)C(C)(C)C)C1=O | 2969.9 | Standard polar | 33892256 | | 1-Methyluric acid,1TBDMS,isomer #2 | CN1C(=O)[NH]C2=C(C1=O)N([Si](C)(C)C(C)(C)C)C(=O)[NH]2 | 2224.3 | Semi standard non polar | 33892256 | | 1-Methyluric acid,1TBDMS,isomer #2 | CN1C(=O)[NH]C2=C(C1=O)N([Si](C)(C)C(C)(C)C)C(=O)[NH]2 | 2428.6 | Standard non polar | 33892256 | | 1-Methyluric acid,1TBDMS,isomer #2 | CN1C(=O)[NH]C2=C(C1=O)N([Si](C)(C)C(C)(C)C)C(=O)[NH]2 | 2903.2 | Standard polar | 33892256 | | 1-Methyluric acid,1TBDMS,isomer #3 | CN1C(=O)[NH]C2=C([NH]C(=O)N2[Si](C)(C)C(C)(C)C)C1=O | 2193.8 | Semi standard non polar | 33892256 | | 1-Methyluric acid,1TBDMS,isomer #3 | CN1C(=O)[NH]C2=C([NH]C(=O)N2[Si](C)(C)C(C)(C)C)C1=O | 2464.5 | Standard non polar | 33892256 | | 1-Methyluric acid,1TBDMS,isomer #3 | CN1C(=O)[NH]C2=C([NH]C(=O)N2[Si](C)(C)C(C)(C)C)C1=O | 2997.6 | Standard polar | 33892256 | | 1-Methyluric acid,2TBDMS,isomer #1 | CN1C(=O)C2=C([NH]C(=O)N2[Si](C)(C)C(C)(C)C)N([Si](C)(C)C(C)(C)C)C1=O | 2407.5 | Semi standard non polar | 33892256 | | 1-Methyluric acid,2TBDMS,isomer #1 | CN1C(=O)C2=C([NH]C(=O)N2[Si](C)(C)C(C)(C)C)N([Si](C)(C)C(C)(C)C)C1=O | 2656.4 | Standard non polar | 33892256 | | 1-Methyluric acid,2TBDMS,isomer #1 | CN1C(=O)C2=C([NH]C(=O)N2[Si](C)(C)C(C)(C)C)N([Si](C)(C)C(C)(C)C)C1=O | 2581.5 | Standard polar | 33892256 | | 1-Methyluric acid,2TBDMS,isomer #2 | CN1C(=O)C2=C(N([Si](C)(C)C(C)(C)C)C(=O)[NH]2)N([Si](C)(C)C(C)(C)C)C1=O | 2403.4 | Semi standard non polar | 33892256 | | 1-Methyluric acid,2TBDMS,isomer #2 | CN1C(=O)C2=C(N([Si](C)(C)C(C)(C)C)C(=O)[NH]2)N([Si](C)(C)C(C)(C)C)C1=O | 2702.2 | Standard non polar | 33892256 | | 1-Methyluric acid,2TBDMS,isomer #2 | CN1C(=O)C2=C(N([Si](C)(C)C(C)(C)C)C(=O)[NH]2)N([Si](C)(C)C(C)(C)C)C1=O | 2634.9 | Standard polar | 33892256 | | 1-Methyluric acid,2TBDMS,isomer #3 | CN1C(=O)[NH]C2=C(C1=O)N([Si](C)(C)C(C)(C)C)C(=O)N2[Si](C)(C)C(C)(C)C | 2391.3 | Semi standard non polar | 33892256 | | 1-Methyluric acid,2TBDMS,isomer #3 | CN1C(=O)[NH]C2=C(C1=O)N([Si](C)(C)C(C)(C)C)C(=O)N2[Si](C)(C)C(C)(C)C | 2689.4 | Standard non polar | 33892256 | | 1-Methyluric acid,2TBDMS,isomer #3 | CN1C(=O)[NH]C2=C(C1=O)N([Si](C)(C)C(C)(C)C)C(=O)N2[Si](C)(C)C(C)(C)C | 2619.4 | Standard polar | 33892256 | | 1-Methyluric acid,3TBDMS,isomer #1 | CN1C(=O)C2=C(N([Si](C)(C)C(C)(C)C)C1=O)N([Si](C)(C)C(C)(C)C)C(=O)N2[Si](C)(C)C(C)(C)C | 2627.1 | Semi standard non polar | 33892256 | | 1-Methyluric acid,3TBDMS,isomer #1 | CN1C(=O)C2=C(N([Si](C)(C)C(C)(C)C)C1=O)N([Si](C)(C)C(C)(C)C)C(=O)N2[Si](C)(C)C(C)(C)C | 2912.7 | Standard non polar | 33892256 | | 1-Methyluric acid,3TBDMS,isomer #1 | CN1C(=O)C2=C(N([Si](C)(C)C(C)(C)C)C1=O)N([Si](C)(C)C(C)(C)C)C(=O)N2[Si](C)(C)C(C)(C)C | 2593.0 | Standard polar | 33892256 |
|
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - 1-Methyluric acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-0far-2900000000-7c88ba8c892ca8263473 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 1-Methyluric acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 1-Methyluric acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Quattro_QQQ 10V, Positive-QTOF (Annotated) | splash10-001i-0900000000-f2e42cd67ce1106bed63 | 2012-07-25 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Quattro_QQQ 25V, Positive-QTOF (Annotated) | splash10-001i-0900000000-9fd2304504c5787dc255 | 2012-07-25 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Quattro_QQQ 40V, Positive-QTOF (Annotated) | splash10-0a4i-9000000000-6069b457c0c038edb8df | 2012-07-25 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Linear Ion Trap , positive-QTOF | splash10-0006-0900000000-f47eba015e7f8d9f21a7 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Orbitrap 4V, negative-QTOF | splash10-001i-0900000000-26da10b11bacabae0d00 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Orbitrap 5V, negative-QTOF | splash10-001i-0900000000-dceee0216914a6ad0349 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Orbitrap 6V, negative-QTOF | splash10-001r-0900000000-cb9d81f2cc810b6a1bf6 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Orbitrap 8V, negative-QTOF | splash10-0019-2900000000-79ddb513e243e4fd1054 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Orbitrap 10V, negative-QTOF | splash10-001a-7900000000-aeb7c72625ab71bbfcb3 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid n/a 12V, negative-QTOF | splash10-000i-0900000000-72f91c756d76768ecc23 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid n/a 12V, negative-QTOF | splash10-004i-9000000000-0ae8d2338f48c17984cd | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid n/a 12V, negative-QTOF | splash10-001i-9400000000-943dd572fd686e87913d | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Orbitrap 1V, negative-QTOF | splash10-03di-0109000000-cf0f38f1f9c792f0404d | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Orbitrap 1V, negative-QTOF | splash10-03di-0309000000-9a8aaf68c144b67fbc6e | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Orbitrap 2V, negative-QTOF | splash10-01q9-0905000000-d40f59a74d4b8cc4e1d6 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Orbitrap 3V, negative-QTOF | splash10-001i-0901000000-265af2cad1ab697d3249 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Orbitrap 3V, negative-QTOF | splash10-001i-0900000000-2b4e4dd83c0bad2269ae | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Orbitrap 4V, negative-QTOF | splash10-001i-0900000000-c37839583f5e4e15cc46 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 1-Methyluric acid Orbitrap 9V, negative-QTOF | splash10-001i-0900000000-6254efb7ddc081f6fd0c | 2020-07-22 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Methyluric acid 10V, Positive-QTOF | splash10-001i-0900000000-9fb7b528d968cab88bd3 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Methyluric acid 20V, Positive-QTOF | splash10-0059-1900000000-eb2d1569b5bdfeef979a | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Methyluric acid 40V, Positive-QTOF | splash10-000w-9300000000-065aa3463c121bf31312 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Methyluric acid 10V, Negative-QTOF | splash10-001i-1900000000-62dd716e79c8fa6f5ee0 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Methyluric acid 20V, Negative-QTOF | splash10-001i-3900000000-bf85a8ad5b7719f36aba | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Methyluric acid 40V, Negative-QTOF | splash10-0006-9100000000-7db60439b29417bbbe0d | 2017-09-01 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 600 MHz, 5%_DMSO, experimental) | 2012-12-05 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-25 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 5%_DMSO, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
|
|---|
| Disease References | | Colorectal cancer |
|---|
- Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP: Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016 Jun 6;4:11. doi: 10.1186/s40170-016-0151-y. eCollection 2016. [PubMed:27275383 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
| | Canavan disease |
|---|
- Tavazzi B, Lazzarino G, Leone P, Amorini AM, Bellia F, Janson CG, Di Pietro V, Ceccarelli L, Donzelli S, Francis JS, Giardina B: Simultaneous high performance liquid chromatographic separation of purines, pyrimidines, N-acetylated amino acids, and dicarboxylic acids for the chemical diagnosis of inborn errors of metabolism. Clin Biochem. 2005 Nov;38(11):997-1008. Epub 2005 Sep 1. [PubMed:16139832 ]
|
|
|---|
| General References | - Knoppert DC, Spino M, Beck R, Thiessen JJ, MacLeod SM: Cystic fibrosis: enhanced theophylline metabolism may be linked to the disease. Clin Pharmacol Ther. 1988 Sep;44(3):254-64. [PubMed:3046811 ]
- Morris GS, Simmonds HA, Davies PM: Use of biological fluids for the rapid diagnosis of potentially lethal inherited disorders of human purine and pyrimidine metabolism. Biomed Chromatogr. 1986 Jun;1(3):109-18. [PubMed:3506820 ]
- Tateishi T, Asoh M, Yamaguchi A, Yoda T, Okano YJ, Koitabashi Y, Kobayashi S: Developmental changes in urinary elimination of theophylline and its metabolites in pediatric patients. Pediatr Res. 1999 Jan;45(1):66-70. [PubMed:9890610 ]
- Nakano K, Assenza SP, Brown PR: Reversed-phase liquid chromatographic investigation of UV-absorbing low-molecular-weight compounds in saliva. J Chromatogr. 1982 Dec 10;233:51-60. [PubMed:7161362 ]
- Grygiel JJ, Wing LM, Farkas J, Birkett DJ: Effects of allopurinol on theophylline metabolism and clearance. Clin Pharmacol Ther. 1979 Nov;26(5):660-7. [PubMed:498708 ]
- Kilbane AJ, Silbart LK, Manis M, Beitins IZ, Weber WW: Human N-acetylation genotype determination with urinary caffeine metabolites. Clin Pharmacol Ther. 1990 Apr;47(4):470-7. [PubMed:2328555 ]
- Safranow K, Machoy Z: Simultaneous determination of 16 purine derivatives in urinary calculi by gradient reversed-phase high-performance liquid chromatography with UV detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2005 May 25;819(2):229-35. [PubMed:15833286 ]
- Safranow K: [Identification and quantitation of purine derivatives in urinary calculi as markers of abnormal purine metabolism by using high-performance liquid chromatography (HPLC)]. Ann Acad Med Stetin. 2000;46:35-49. [PubMed:11712316 ]
- Caubet MS, Comte B, Brazier JL: Determination of urinary 13C-caffeine metabolites by liquid chromatography-mass spectrometry: the use of metabolic ratios to assess CYP1A2 activity. J Pharm Biomed Anal. 2004 Feb 4;34(2):379-89. [PubMed:15013152 ]
- Miller CA, Slusher LB, Vesell ES: Polymorphism of theophylline metabolism in man. J Clin Invest. 1985 May;75(5):1415-25. [PubMed:4039734 ]
|
|---|