| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2006-08-13 07:46:56 UTC |
|---|
| Update Date | 2022-03-07 02:49:20 UTC |
|---|
| HMDB ID | HMDB0003938 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | (S)-Hydroxydecanoyl-CoA |
|---|
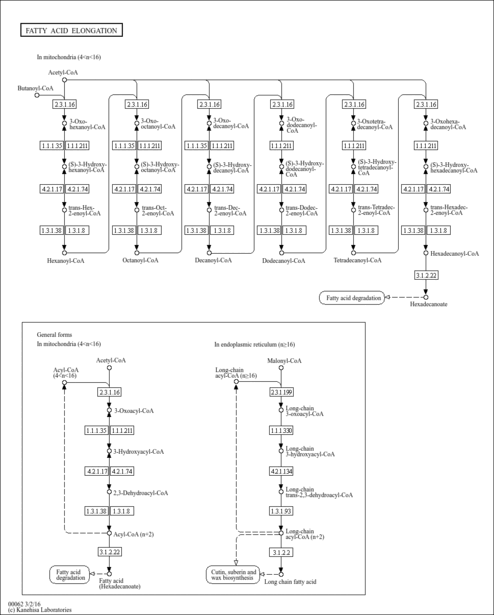
| Description | (s)-hydroxydecanoyl-coa, also known as S-(3-hydroxydecanoate) CoA or 3S-hydroxy-decanoyl-CoA is an acyl-CoA or acyl-coenzyme A. More specifically, it is a 3-hydroxydecanoic acid thioester of coenzyme A. (s)-hydroxydecanoyl-coa is an acyl-CoA with 10 fatty acid group as the acyl moiety attached to coenzyme A. Coenzyme A was discovered in 1946 by Fritz Lipmann (Journal of Biological Chemistry (1946) 162 (3): 743–744) and its structure was determined in the early 1950s at the Lister Institute in London. Coenzyme A is a complex, thiol-containing molecule that is naturally synthesized from pantothenate (vitamin B5), which is found in various foods such as meat, vegetables, cereal grains, legumes, eggs, and milk. More specifically, coenzyme A (CoASH or CoA) consists of a beta-mercaptoethylamine group linked to the vitamin pantothenic acid (B5) through an amide linkage and 3'-phosphorylated ADP. Coenzyme A is synthesized in a five-step process that requires four molecules of ATP, pantothenate and cysteine. It is believed that there are more than 1100 types of acyl-CoA’s in the human body, which also corresponds to the number of acylcarnitines in the human body. Acyl-CoAs exists in all living species, ranging from bacteria to plants to humans. The general role of acyl-CoA’s is to assist in transferring fatty acids from the cytoplasm to mitochondria. This process facilitates the production of fatty acids in cells, which are essential in cell membrane structure. Acyl-CoA's are also susceptible to beta oxidation, forming, ultimately, acetyl-CoA. Acetyl-CoA can enter the citric acid cycle, eventually forming several equivalents of ATP. In this way, fats are converted to ATP -- or biochemical energy. Acyl-CoAs can be classified into 9 different categories depending on the size of their acyl-group: 1) short-chain acyl-CoAs; 2) medium-chain acyl-CoAs; 3) long-chain acyl-CoAs; and 4) very long-chain acyl-CoAs; 5) hydroxy acyl-CoAs; 6) branched chain acyl-CoAs; 7) unsaturated acyl-CoAs; 8) dicarboxylic acyl-CoAs and 9) miscellaneous acyl-CoAs. Short-chain acyl-CoAs have acyl-groups with two to four carbons (C2-C4), medium-chain acyl-CoAs have acyl-groups with five to eleven carbons (C5-C11), long-chain acyl-CoAs have acyl-groups with twelve to twenty carbons (C12-C20) while very long-chain acyl-CoAs have acyl groups with more than 20 carbons. (s)-hydroxydecanoyl-coa is therefore classified as a medium chain acyl-CoA. The oxidative degradation of fatty acids is a two-step process, catalyzed by acyl-CoA synthetase/synthase. Fatty acids are first converted to their acyl phosphate, the precursor to acyl-CoA. The latter conversion is mediated by acyl-CoA synthase. Three types of acyl-CoA synthases are employed, depending on the chain length of the fatty acid. (s)-hydroxydecanoyl-coa, being a medium chain acyl-CoA is a substrate for medium chain acyl-CoA synthase. The second step of fatty acid degradation is beta oxidation. Beta oxidation occurs in mitochondria and, in the case of very long chain acyl-CoAs, the peroxisome. After its formation in the cytosol, (S)-Hydroxydecanoyl-CoA is transported into the mitochondria, the locus of beta oxidation. Transport of (S)-Hydroxydecanoyl-CoA into the mitochondria requires carnitine palmitoyltransferase 1 (CPT1), which converts (S)-Hydroxydecanoyl-CoA into 3-Hydroxydecanoylcarnitine, which gets transported into the mitochondrial matrix. Once in the matrix, 3-Hydroxydecanoylcarnitine is converted back to (S)-Hydroxydecanoyl-CoA by CPT2, whereupon beta-oxidation can begin. Beta oxidation of (S)-Hydroxydecanoyl-CoA occurs in four steps. First, since (S)-Hydroxydecanoyl-CoA is a medium chain acyl-CoA it is the substrate for a medium chain acyl-CoA dehydrogenase, which catalyzes dehydrogenation of (S)-Hydroxydecanoyl-CoA, creating a double bond between the alpha and beta carbons. FAD is the hydrogen acceptor, yielding FADH2. Second, Enoyl-CoA hydrase catalyzes the addition of water across the newly formed double bond to make an alcohol. Third, 3-hydroxyacyl-CoA dehydrogenase oxidizes the alcohol group to a ketone and NADH is produced from NAD+. Finally, Thiolase cleaves between the alpha carbon and ketone to release one molecule of acetyl-CoA and a new acyl-CoA which is now 2 carbons shorter. This four-step process repeats until (S)-Hydroxydecanoyl-CoA has had all its carbons removed from the chain, leaving only acetyl-CoA. Beta oxidation, as well as alpha-oxidation, also occurs in the peroxisome. The peroxisome handles beta oxidation of fatty acids that have more than 20 carbons in their chain because the peroxisome contains very-long-chain Acyl-CoA synthetases and dehydrogenases. The heart primarily metabolizes fat for energy and Acyl-CoA metabolism has been identified as a critical molecule in early-stage heart muscle pump failure. Cellular acyl-CoA content also correlates with insulin resistance, suggesting that it can mediate lipotoxicity in non-adipose tissues. Acyl-CoA: diacylglycerol acyltransferase (DGAT) plays an important role in energy metabolism on account of key enzyme in triglyceride biosynthesis. The study of acyl-CoAs is an active area of research and it is likely that many novel acyl-CoAs will be discovered in the coming years. It is also likely that many novel roles in health and disease will be uncovered for these molecules. |
|---|
| Structure | CCCCCCC[C@H](O)CC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H](C(O)[C@H]1OP(O)(O)=O)N1C=NC2=C(N)N=CN=C12 InChI=1S/C31H54N7O18P3S/c1-4-5-6-7-8-9-19(39)14-22(41)60-13-12-33-21(40)10-11-34-29(44)26(43)31(2,3)16-53-59(50,51)56-58(48,49)52-15-20-25(55-57(45,46)47)24(42)30(54-20)38-18-37-23-27(32)35-17-36-28(23)38/h17-20,24-26,30,39,42-43H,4-16H2,1-3H3,(H,33,40)(H,34,44)(H,48,49)(H,50,51)(H2,32,35,36)(H2,45,46,47)/t19-,20+,24?,25-,26-,30+/m0/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| (S)-3-Hydroxydecanoyl-CoA | HMDB | | (S)-3-Hydroxydecanoyl-coenzyme A | HMDB | | 3-Hydroxydecanoyl-CoA | HMDB | | 3-Hydroxydecanoyl-coenzyme A | HMDB | | 3S-Hydroxy-decanoyl-CoA | HMDB | | 3S-Hydroxy-decanoyl-coenzyme A | HMDB | | b-Hydroxydecanoyl coenzyme A | HMDB | | beta-Hydroxydecanoyl coenzyme A | HMDB | | DL-b-Hydroxydecanoyl coenzyme A | HMDB | | DL-beta-Hydroxydecanoyl coenzyme A | HMDB | | S-(3-Hydroxydecanoate | HMDB | | S-(3-Hydroxydecanoate) CoA | HMDB | | S-(3-Hydroxydecanoate) coenzyme A | HMDB | | S-(3-Hydroxydecanoate)CoA | HMDB | | S-(3-Hydroxydecanoate)coenzyme A | HMDB | | S-(3-Hydroxydecanoic acid | HMDB | | 3-Hydroxydecanoyl-coenzyme A, (R)-isomer | HMDB |
|
|---|
| Chemical Formula | C31H54N7O18P3S |
|---|
| Average Molecular Weight | 937.783 |
|---|
| Monoisotopic Molecular Weight | 937.245888185 |
|---|
| IUPAC Name | {[(2R,3R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-({[hydroxy({hydroxy[(3R)-3-hydroxy-3-({2-[(2-{[(3S)-3-hydroxydecanoyl]sulfanyl}ethyl)carbamoyl]ethyl}carbamoyl)-2,2-dimethylpropoxy]phosphoryl}oxy)phosphoryl]oxy}methyl)oxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | (S)-3-hydroxydecanoyl-coa |
|---|
| CAS Registry Number | 6245-70-1 |
|---|
| SMILES | CCCCCCC[C@H](O)CC(=O)SCCNC(=O)CCNC(=O)[C@H](O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H](C(O)[C@H]1OP(O)(O)=O)N1C=NC2=C(N)N=CN=C12 |
|---|
| InChI Identifier | InChI=1S/C31H54N7O18P3S/c1-4-5-6-7-8-9-19(39)14-22(41)60-13-12-33-21(40)10-11-34-29(44)26(43)31(2,3)16-53-59(50,51)56-58(48,49)52-15-20-25(55-57(45,46)47)24(42)30(54-20)38-18-37-23-27(32)35-17-36-28(23)38/h17-20,24-26,30,39,42-43H,4-16H2,1-3H3,(H,33,40)(H,34,44)(H,48,49)(H,50,51)(H2,32,35,36)(H2,45,46,47)/t19-,20+,24?,25-,26-,30+/m0/s1 |
|---|
| InChI Key | HIVSMYZAMUNFKZ-WQUYVQPTSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as (s)-3-hydroxyacyl coas. These are organic compounds containing a (S)-3-hydroxyl acylated coenzyme A derivative. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | (S)-3-hydroxyacyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Pyrimidine
- Alkyl phosphate
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Azole
- Heteroaromatic compound
- Carbothioic s-ester
- Secondary alcohol
- Thiocarboxylic acid ester
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Organosulfur compound
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organic nitrogen compound
- Primary amine
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | -1.219 | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 4.78 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 11.2307 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 9.21 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 501.0 seconds | 40023050 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 1719.0 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 150.0 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 132.9 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 167.0 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 110.0 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 411.0 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 494.8 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 1061.0 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 754.9 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 448.2 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 764.0 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 261.5 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 254.8 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 447.7 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 262.4 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 312.8 seconds | 40023050 |
Predicted Kovats Retention IndicesNot Available |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| MS | Mass Spectrum (Electron Ionization) | Not Available | 2022-08-06 | Not Available | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (S)-Hydroxydecanoyl-CoA 10V, Positive-QTOF | splash10-000i-1912101104-e68576ec41a01a4493b0 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (S)-Hydroxydecanoyl-CoA 20V, Positive-QTOF | splash10-000i-0931400000-9d5e56a46993e0feaed8 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (S)-Hydroxydecanoyl-CoA 40V, Positive-QTOF | splash10-000i-1900101000-3bf523a37d679f13333f | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (S)-Hydroxydecanoyl-CoA 10V, Negative-QTOF | splash10-017r-2911030304-9f3643027fe9f8830513 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (S)-Hydroxydecanoyl-CoA 20V, Negative-QTOF | splash10-001i-2911110001-b12adbca1de54d3563d8 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (S)-Hydroxydecanoyl-CoA 40V, Negative-QTOF | splash10-057i-6900000000-25ae0704fa36fc9ea146 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (S)-Hydroxydecanoyl-CoA 10V, Negative-QTOF | splash10-000i-0000000009-80e02b0bfbb587a9f439 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (S)-Hydroxydecanoyl-CoA 20V, Negative-QTOF | splash10-0170-4200201229-44afe866118bc7aaffa3 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (S)-Hydroxydecanoyl-CoA 40V, Negative-QTOF | splash10-00ou-3404501933-d11a9fe0cd5738a22791 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (S)-Hydroxydecanoyl-CoA 10V, Positive-QTOF | splash10-00dr-0000000009-1425c44ee49950fbde61 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (S)-Hydroxydecanoyl-CoA 20V, Positive-QTOF | splash10-03mr-0000100195-67ca18096ef07994d794 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (S)-Hydroxydecanoyl-CoA 40V, Positive-QTOF | splash10-001i-0012900000-11900216d077fbcceb67 | 2021-09-23 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum |
|
|---|