| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2006-08-13 07:55:12 UTC |
|---|
| Update Date | 2022-03-07 02:49:20 UTC |
|---|
| HMDB ID | HMDB0003949 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | (2E)-Octenoyl-CoA |
|---|
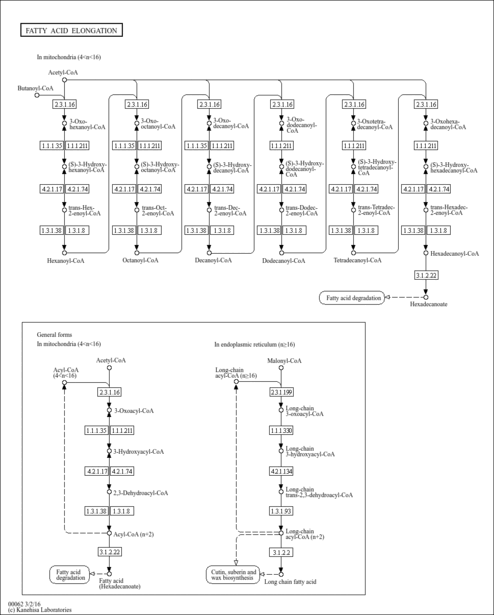
| Description | (2E)-Octenoyl-CoA is the main metabolite produced in medium-chain acyl-CoA dehydrogenase (EC 1.3.99.3, MCAD) deficiency; however the product of the enzymatic reaction is not directly detected in several methods for screening of inborn errors of fatty acid oxidation. In order to aid the timely follow-up of screening results that suggest abnormalities in MCAD, rapid and simple confirmatory tests for the enzyme activity and/or gene mutation analysis should be available. Medium-chain fatty acyl-CoA dehydrogenase (MCAD) catalyzes the conversion of different chain length fatty acyl- CoAs into their corresponding trans-enoyl-CoA moieties via two consecutive sequences of steps. The first step involves the concerted abstraction of a proton and a hydride ion from the a- and 8-carbon chains of the fatty acyl-CoA substrates, concomitant with the reduction of the enzyme (E)-bound FAD to FADH2. The reoxidation of EFADH2, to propagate further rounds of catalysis, is accomplished via transfer of electrons to a variety of organic electron acceptors; the natural electron acceptor for this process, under physiological conditions, is the electron-transferring flavoprotein. Of the different chain length fatty acyl-CoA substrates, octanoyl-CoA/octenoyl-CoA have been known as the most efficient (and physiological) substrates for the medium-chain fatty acyl-CoA dehydrogenase (MCAD)-catalyzed reaction. (PMID: 16046200 , 1390638 , 8038175 ). |
|---|
| Structure | CCCCC\C=C\C(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N InChI=1S/C29H48N7O17P3S/c1-4-5-6-7-8-9-20(38)57-13-12-31-19(37)10-11-32-27(41)24(40)29(2,3)15-50-56(47,48)53-55(45,46)49-14-18-23(52-54(42,43)44)22(39)28(51-18)36-17-35-21-25(30)33-16-34-26(21)36/h8-9,16-18,22-24,28,39-40H,4-7,10-15H2,1-3H3,(H,31,37)(H,32,41)(H,45,46)(H,47,48)(H2,30,33,34)(H2,42,43,44)/b9-8+/t18-,22-,23-,24?,28-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| (e)-S-2-Octenoate | HMDB | | (e)-S-2-Octenoate CoA | HMDB | | (e)-S-2-Octenoate coenzyme A | HMDB | | (e)-S-2-Octenoic acid | HMDB | | 2,3-trans-Octenoyl coenzyme A | HMDB | | Oct-2-trans-enoyl-CoA | HMDB | | Oct-2-trans-enoyl-coenzyme A | HMDB | | Oct-trans-2-enoyl coenzyme A | HMDB | | S-(2E)-2-Octenoate | HMDB | | S-(2E)-2-Octenoate CoA | HMDB | | S-(2E)-2-Octenoate coenzyme A | HMDB | | S-(2E)-2-Octenoic acid | HMDB | | trans-D2,3-Octenoyl-CoA | HMDB | | trans-D2,3-Octenoyl-coenzyme A | HMDB | | trans-Oct-2-enoyl-CoA | HMDB | | trans-Oct-2-enoyl-coenzyme A | HMDB |
|
|---|
| Chemical Formula | C29H48N7O17P3S |
|---|
| Average Molecular Weight | 891.714 |
|---|
| Monoisotopic Molecular Weight | 891.204023371 |
|---|
| IUPAC Name | {[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-({[hydroxy({[hydroxy(3-hydroxy-2,2-dimethyl-3-{[2-({2-[(2E)-oct-2-enoylsulfanyl]ethyl}carbamoyl)ethyl]carbamoyl}propoxy)phosphoryl]oxy})phosphoryl]oxy}methyl)oxolan-3-yl]oxy}phosphonic acid |
|---|
| Traditional Name | [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-2-[({hydroxy[hydroxy(3-hydroxy-2,2-dimethyl-3-{[2-({2-[(2E)-oct-2-enoylsulfanyl]ethyl}carbamoyl)ethyl]carbamoyl}propoxy)phosphoryl]oxyphosphoryl}oxy)methyl]oxolan-3-yl]oxyphosphonic acid |
|---|
| CAS Registry Number | 10018-94-7 |
|---|
| SMILES | CCCCC\C=C\C(=O)SCCNC(=O)CCNC(=O)C(O)C(C)(C)COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C29H48N7O17P3S/c1-4-5-6-7-8-9-20(38)57-13-12-31-19(37)10-11-32-27(41)24(40)29(2,3)15-50-56(47,48)53-55(45,46)49-14-18-23(52-54(42,43)44)22(39)28(51-18)36-17-35-21-25(30)33-16-34-26(21)36/h8-9,16-18,22-24,28,39-40H,4-7,10-15H2,1-3H3,(H,31,37)(H,32,41)(H,45,46)(H,47,48)(H2,30,33,34)(H2,42,43,44)/b9-8+/t18-,22-,23-,24?,28-/m1/s1 |
|---|
| InChI Key | CPSDNAXXKWVYIY-DPSCIZRPSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as medium-chain 2-enoyl coas. These are organic compounds containing a coenzyme A substructure linked to a medium-chain 2-enoyl chain of 5 to 12 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | Medium-chain 2-enoyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Pyrimidine
- Alkyl phosphate
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Azole
- Heteroaromatic compound
- Carbothioic s-ester
- Secondary alcohol
- Thiocarboxylic acid ester
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Organosulfur compound
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organic nitrogen compound
- Primary amine
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | -1.281 | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 4.4 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 11.6646 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 9.09 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 448.5 seconds | 40023050 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 1892.1 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 156.2 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 124.7 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 163.5 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 108.4 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 431.8 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 525.3 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 918.9 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 803.0 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 470.1 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 768.3 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 304.1 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 244.1 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 416.6 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 239.3 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 190.5 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatized |
|---|
| MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (2E)-Octenoyl-CoA 10V, Positive-QTOF | splash10-000i-1912000120-6a4c400b3152685ddc42 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (2E)-Octenoyl-CoA 20V, Positive-QTOF | splash10-000i-0933000000-6bfba9e629e0c41c1c83 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (2E)-Octenoyl-CoA 40V, Positive-QTOF | splash10-000i-1911000000-7bd78cc2705414ac12aa | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (2E)-Octenoyl-CoA 10V, Negative-QTOF | splash10-00gl-3911030340-0a6b8d53ed19f026db3f | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (2E)-Octenoyl-CoA 20V, Negative-QTOF | splash10-00ai-3910010010-8cc155429b1ae945edc0 | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (2E)-Octenoyl-CoA 40V, Negative-QTOF | splash10-057i-6900000000-8852cd72aadbe50cd70b | 2015-09-15 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (2E)-Octenoyl-CoA 10V, Positive-QTOF | splash10-0006-0000000190-ffcd2eab73ff60157945 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (2E)-Octenoyl-CoA 20V, Positive-QTOF | splash10-000i-1910101580-d4dfe3fd333b1bef04a9 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (2E)-Octenoyl-CoA 40V, Positive-QTOF | splash10-000i-0029000000-94de84f8ae35a8bb2b05 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (2E)-Octenoyl-CoA 10V, Negative-QTOF | splash10-0006-0000000090-15ef43bffda7c547e777 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (2E)-Octenoyl-CoA 20V, Negative-QTOF | splash10-009f-5300303790-609e6396a205775b94b1 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - (2E)-Octenoyl-CoA 40V, Negative-QTOF | splash10-014r-4103402920-b4cc1e40b5e853471683 | 2021-09-22 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-24 | Wishart Lab | View Spectrum |
|
|---|
| General References | - Tajima G, Sakura N, Yofune H, Nishimura Y, Ono H, Hasegawa Y, Hata I, Kimura M, Yamaguchi S, Shigematsu Y, Kobayashi M: Enzymatic diagnosis of medium-chain acyl-CoA dehydrogenase deficiency by detecting 2-octenoyl-CoA production using high-performance liquid chromatography: a practical confirmatory test for tandem mass spectrometry newborn screening in Japan. J Chromatogr B Analyt Technol Biomed Life Sci. 2005 Sep 5;823(2):122-30. [PubMed:16046200 ]
- Cummings JG, Lau SM, Powell PJ, Thorpe C: Reductive half-reaction in medium-chain acyl-CoA dehydrogenase: modulation of internal equilibrium by carboxymethylation of a specific methionine residue. Biochemistry. 1992 Sep 15;31(36):8523-9. [PubMed:1390638 ]
- Kumar NR, Srivastava DK: Reductive half-reaction of medium-chain fatty acyl-CoA dehydrogenase utilizing octanoyl-CoA/octenoyl-CoA as a physiological substrate/product pair: similarity in the microscopic pathways of octanoyl-CoA oxidation and octenoyl-CoA binding. Biochemistry. 1994 Jul 26;33(29):8833-41. [PubMed:8038175 ]
|
|---|