| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2023-05-30 20:55:54 UTC |
|---|
| HMDB ID | HMDB0000011 |
|---|
| Secondary Accession Numbers | - HMDB0000357
- HMDB00011
- HMDB00357
|
|---|
| Metabolite Identification |
|---|
| Common Name | 3-Hydroxybutyric acid |
|---|
| Description | 3-Hydroxybutyric acid (CAS: 300-85-6), also known as beta-hydroxybutanoic acid, is a typical partial-degradation product of branched-chain amino acids (primarily valine) released from muscle for hepatic and renal gluconeogenesis. This acid is metabolized by 3-hydroxybutyrate dehydrogenase (catalyzes the oxidation of 3-hydroxybutyrate to form acetoacetate, using NAD+ as an electron acceptor). The enzyme functions in nervous tissues and muscles, enabling the use of circulating hydroxybutyrate as a fuel. In the liver mitochondrial matrix, the enzyme can also catalyze the reverse reaction, a step in ketogenesis. 3-Hydroxybutyric acid is a chiral compound having two enantiomers, D-3-hydroxybutyric acid and L-3-hydroxybutyric acid, and is a ketone body. Like the other ketone bodies (acetoacetate and acetone), levels of 3-hydroxybutyrate in blood and urine are raised in ketosis. In humans, 3-hydroxybutyrate is synthesized in the liver from acetyl-CoA and can be used as an energy source by the brain when blood glucose is low. Blood levels of 3-hydroxybutyric acid levels may be monitored in diabetic patients to look for diabetic ketoacidosis. Persistent mild hyperketonemia is a common finding in newborns. Ketone bodies serve as an indispensable source of energy for extrahepatic tissues, especially the brain and lung of developing mammals. Another important function of ketone bodies is to provide acetoacetyl-CoA and acetyl-CoA for the synthesis of cholesterol, fatty acids, and complex lipids. During the early postnatal period, acetoacetate (AcAc) and beta-hydroxybutyrate are preferred over glucose as substrates for the synthesis of phospholipids and sphingolipids in accord with requirements for brain growth and myelination. Thus, during the first two weeks of postnatal development, when the accumulation of cholesterol and phospholipids accelerates, the proportion of ketone bodies incorporated into these lipids increases. On the other hand, an increased proportion of ketone bodies is utilized for cerebroside synthesis during the period of active myelination. In the lung, AcAc serves better than glucose as a precursor for the synthesis of lung phospholipids. The synthesized lipids, particularly dipalmitoylphosphatidylcholine, are incorporated into surfactant, and thus have a potential role in supplying adequate surfactant lipids to maintain lung function during the early days of life (PMID: 3884391 ). 3-Hydroxybutyric acid is found to be associated with fumarase deficiency and medium-chain acyl-CoA dehydrogenase deficiency, which are inborn errors of metabolism. 3-Hydroxybutyric acid is a metabolite of Alcaligenes and can be produced from plastic metabolization or incorporated into polymers, depending on the species (PMID: 7646009 , 18615882 ). |
|---|
| Structure | InChI=1S/C4H8O3/c1-3(5)2-4(6)7/h3,5H,2H2,1H3,(H,6,7)/t3-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| (R)-(-)-beta-Hydroxybutyric acid | ChEBI | | (R)-3-Hydroxybutanoic acid | ChEBI | | 3-D-Hydroxybutyric acid | ChEBI | | D-3-Hydroxybutyric acid | ChEBI | | D-beta-Hydroxybutyric acid | Kegg | | (R)-(-)-b-Hydroxybutyrate | Generator | | (R)-(-)-b-Hydroxybutyric acid | Generator | | (R)-(-)-beta-Hydroxybutyrate | Generator | | (R)-(-)-Β-hydroxybutyrate | Generator | | (R)-(-)-Β-hydroxybutyric acid | Generator | | (R)-3-Hydroxybutanoate | Generator | | 3-D-Hydroxybutyrate | Generator | | D-3-Hydroxybutyrate | Generator | | D-b-Hydroxybutyrate | Generator | | D-b-Hydroxybutyric acid | Generator | | D-beta-Hydroxybutyrate | Generator | | D-Β-hydroxybutyrate | Generator | | D-Β-hydroxybutyric acid | Generator | | (R)-3-Hydroxybutyrate | Generator | | 3-delta-Hydroxybutyrate | HMDB | | 3-delta-Hydroxybutyric acid | HMDB | | BHIB | HMDB | | D-(-)-3-Hydroxybutyrate | HMDB | | delta-(-)-3-Hydroxybutyrate | HMDB | | delta-3-Hydroxybutyrate | HMDB | | delta-3-Hydroxybutyric acid | HMDB | | delta-beta-Hydroxybutyrate | HMDB | | 3R-Hydroxy-butanoate | HMDB | | (R)-3-Hydroxybutyric acid | KEGG | | (-)-3-Hydroxy-n-butyric acid | HMDB | | (-)-3-Hydroxybutyric acid | HMDB | | (3R)-3-Hydroxybutanoic acid | HMDB | | (3R)-3-Hydroxybutyric acid | HMDB | | (3R)-Hydroxybutyrate | HMDB | | (R)-(-)-3-Hydroxybutyric acid | HMDB | | (R)-beta-Hydroxybutanoic acid | HMDB | | (R)-beta-Hydroxybutyric acid | HMDB | | (R)-β-Hydroxybutanoic acid | HMDB | | (R)-β-Hydroxybutyric acid | HMDB | | 3-Hydroxy-n-butyric acid | HMDB | | 3-Hydroxybutanoic acid | HMDB | | 3-Hydroxybutyric acid | HMDB | | 3R-Hydroxybutanoic acid | HMDB | | D-(-)-3-Hydroxybutanoic acid | HMDB | | D-(-)-3-Hydroxybutyric acid | HMDB | | D-(-)-beta-Hydroxybutyric acid | HMDB | | D-(-)-β-Hydroxybutyric acid | HMDB | | beta-Hydroxy-n-butyric acid | HMDB | | beta-Hydroxybutanoic acid | HMDB | | beta-Hydroxybutyric acid | HMDB | | β-Hydroxy-n-butyric acid | HMDB | | β-Hydroxybutanoic acid | HMDB | | β-Hydroxybutyric acid | HMDB |
|
|---|
| Chemical Formula | C4H8O3 |
|---|
| Average Molecular Weight | 104.1045 |
|---|
| Monoisotopic Molecular Weight | 104.047344122 |
|---|
| IUPAC Name | (3R)-3-hydroxybutanoic acid |
|---|
| Traditional Name | (-)-3-hydroxybutyric acid |
|---|
| CAS Registry Number | 625-72-9 |
|---|
| SMILES | C[C@@H](O)CC(O)=O |
|---|
| InChI Identifier | InChI=1S/C4H8O3/c1-3(5)2-4(6)7/h3,5H,2H2,1H3,(H,6,7)/t3-/m1/s1 |
|---|
| InChI Key | WHBMMWSBFZVSSR-GSVOUGTGSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as beta hydroxy acids and derivatives. Beta hydroxy acids and derivatives are compounds containing a carboxylic acid substituted with a hydroxyl group on the C3 carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Hydroxy acids and derivatives |
|---|
| Sub Class | Beta hydroxy acids and derivatives |
|---|
| Direct Parent | Beta hydroxy acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Short-chain hydroxy acid
- Beta-hydroxy acid
- Fatty acid
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 49 - 50 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| 3-Hydroxybutyric acid,1TMS,isomer #1 | C[C@H](CC(=O)O)O[Si](C)(C)C | 1074.4 | Semi standard non polar | 33892256 | | 3-Hydroxybutyric acid,1TMS,isomer #2 | C[C@@H](O)CC(=O)O[Si](C)(C)C | 1018.1 | Semi standard non polar | 33892256 | | 3-Hydroxybutyric acid,2TMS,isomer #1 | C[C@H](CC(=O)O[Si](C)(C)C)O[Si](C)(C)C | 1168.2 | Semi standard non polar | 33892256 | | 3-Hydroxybutyric acid,1TBDMS,isomer #1 | C[C@H](CC(=O)O)O[Si](C)(C)C(C)(C)C | 1313.6 | Semi standard non polar | 33892256 | | 3-Hydroxybutyric acid,1TBDMS,isomer #2 | C[C@@H](O)CC(=O)O[Si](C)(C)C(C)(C)C | 1235.9 | Semi standard non polar | 33892256 | | 3-Hydroxybutyric acid,2TBDMS,isomer #1 | C[C@H](CC(=O)O[Si](C)(C)C(C)(C)C)O[Si](C)(C)C(C)(C)C | 1603.0 | Semi standard non polar | 33892256 |
|
|---|
| Spectra |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - 3-Hydroxybutyric acid EI-B (Non-derivatized) | splash10-0002-0900000000-7410b547b9d02c6326c7 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 3-Hydroxybutyric acid EI-B (Non-derivatized) | splash10-0002-0900000000-7410b547b9d02c6326c7 | 2018-05-18 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 3-Hydroxybutyric acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-0007-9000000000-5f169537ace358b06fd0 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 3-Hydroxybutyric acid GC-MS (2 TMS) - 70eV, Positive | splash10-01ei-9710000000-2652bd46b41e50defdb0 | 2017-10-06 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 3-Hydroxybutyric acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 3-Hydroxybutyric acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 3-Hydroxybutyric acid GC-MS (TMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 3-Hydroxybutyric acid GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 3-Hydroxybutyric acid GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 3-Hydroxybutyric acid GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 3-Hydroxybutyric acid GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | MS | Mass Spectrum (Electron Ionization) | splash10-0007-9000000000-7fc414806153ebd8822c | 2020-03-25 | Not Available | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid Quattro_QQQ 10V, Negative-QTOF (Annotated) | splash10-0a4i-9200000000-4156904e7472b5e97249 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid Quattro_QQQ 25V, Negative-QTOF (Annotated) | splash10-0a4i-9300000000-505ae46abb0c49c78b1e | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid Quattro_QQQ 40V, Negative-QTOF (Annotated) | splash10-0zfr-9600000000-dfed69c37c1a4d794440 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative-QTOF | splash10-0udi-1900000000-b34932b6f2ee5bca9697 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative-QTOF | splash10-0a4i-9100000000-cdbb3586477ed705b4d7 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative-QTOF | splash10-0a4i-9000000000-022ab74e163dd752d054 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative-QTOF | splash10-0006-9000000000-18a18150fcc1dfee58cf | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative-QTOF | splash10-0006-9000000000-e8d8003bcda8aa818d4e | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid LC-ESI-QQ , negative-QTOF | splash10-0udi-1900000000-b34932b6f2ee5bca9697 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid LC-ESI-QQ , negative-QTOF | splash10-0a4i-9100000000-cdbb3586477ed705b4d7 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid LC-ESI-QQ , negative-QTOF | splash10-0a4i-9000000000-022ab74e163dd752d054 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid LC-ESI-QQ , negative-QTOF | splash10-0006-9000000000-187f6a4024ca6e901188 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid LC-ESI-QQ , negative-QTOF | splash10-0006-9000000000-e8d8003bcda8aa818d4e | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid , negative-QTOF | splash10-0a4i-9100000000-79f60e90c0ce32d976b3 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid 35V, Negative-QTOF | splash10-0a4i-9200000000-b3917779cc00018df7d0 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid 10V, Positive-QTOF | splash10-002e-9000000000-d601a5284802b5564ac8 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid 40V, Positive-QTOF | splash10-0udi-9000000000-128801e4b0f2e4d5344b | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid 35V, Negative-QTOF | splash10-0a4i-9100000000-81d99c8d5304bd26a0cd | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - 3-Hydroxybutyric acid 35V, Negative-QTOF | splash10-0a4i-9200000000-fc7c6bb126f183e27ff0 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxybutyric acid 10V, Positive-QTOF | splash10-00kr-9100000000-889e2968ad4fd52cdd92 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxybutyric acid 20V, Positive-QTOF | splash10-05n0-9000000000-6c660170b8f4aa6bcd02 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxybutyric acid 40V, Positive-QTOF | splash10-0006-9000000000-c39869c905b7e93b7f97 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxybutyric acid 10V, Negative-QTOF | splash10-0zfr-9800000000-532ea53160d6ca2efcaa | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxybutyric acid 20V, Negative-QTOF | splash10-0pbi-9200000000-b2c59fd56b1a1d713ea9 | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 3-Hydroxybutyric acid 40V, Negative-QTOF | splash10-0a4l-9000000000-29e8d104108ac71cd640 | 2017-09-01 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
|
|---|
| Biological Properties |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Mitochondria
|
|---|
| Biospecimen Locations | - Blood
- Cellular Cytoplasm
- Cerebrospinal Fluid (CSF)
- Feces
- Saliva
- Urine
|
|---|
| Tissue Locations | Not Available |
|---|
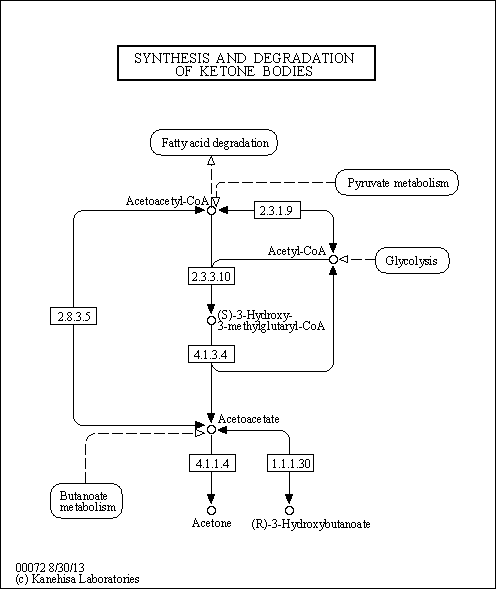
| Pathways | |
|---|
| Normal Concentrations |
|---|
| |
| Blood | Detected and Quantified | 60.0 +/- 20.0 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 40.0 +/- 10.0 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 323.0 +/- 29.0 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 36 +/- 20 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 86 +/- 53 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 38 +/- 31 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | <500 uM | Infant (0-1 year old) | Not Specified | Normal | | details | | Blood | Detected and Quantified | 260-5040 uM | Infant (0-1 year old) | Not Specified | Normal | | details | | Blood | Detected and Quantified | 50.0 (0.0-100.0) uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 46.5 +/- 10.1 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | <600 uM | Not Specified | Not Specified | Normal | | details | | Blood | Detected and Quantified | <150 uM | Not Specified | Not Specified | Normal | | details | | Blood | Detected and Quantified | 100.0 +/- 90.0 uM | Adult (>18 years old) | Female | Normal | | details | | Blood | Detected and Quantified | 565.0 +/- 255.0 uM | Newborn (0-30 days old) | Both | Normal | | details | | Blood | Detected and Quantified | 36.0 (13.0-95.0) uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 80-3600 uM | Not Specified | Not Specified | Normal | | details | | Blood | Detected and Quantified | 64.88(60.4) uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 200 +/- 120 uM | Children (1-13 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 170 +/- 110 uM | Children (1-13 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 10.60-143.2 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 500-3000 uM | Children (1-13 years old) | Male | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 76.9 +/- 66.3 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 180.0 (22.0-700.0) uM | Adult (>18 years old) | Both | Normal | | details | | Cellular Cytoplasm | Detected and Quantified | 1300 (1200-1400) uM | Adult (>18 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 34 +/- 31 uM | Adult (>18 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 50.0 (4.0 - 96.0) uM | Adult (>18 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 286 (207-365) uM | Adult (>18 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 46.4 +/- 23.9 uM | Adult (>18 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 48.0 +/- 48.0 uM | Adult (>18 years old) | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected and Quantified | 22.0932 +/- 9.606 nmol/g wet feces | Not Specified | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Not Specified | Not Specified | Normal | | details | | Saliva | Detected and Quantified | 5.00 +/- 4.25 uM | Adult (>18 years old) | Not Specified | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Not Specified | Normal | | details | | Saliva | Detected and Quantified | 2.64 +/- 1.83 uM | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Saliva | Detected and Quantified | 3.22 +/- 2.77 uM | Adult (>18 years old) | Not Specified | Normal | | details | | Saliva | Detected and Quantified | 3.88 +/- 4.26 uM | Adult (>18 years old) | Both | Normal | | details | | Saliva | Detected and Quantified | 16.44 +/- 19.52 uM | Adult (>18 years old) | Female | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Male | Normal | | details | | Saliva | Detected and Quantified | 4.71 +/- 7.21 uM | Adult (>18 years old) | Female | Normal | | details | | Urine | Detected and Quantified | 0.5-9.8 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | <99 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 1.4-2.7 umol/mmol creatinine | Adult (>18 years old) | Female | Normal | | details | | Urine | Detected and Quantified | <54.301 umol/mmol creatinine | Infant (0-1 year old) | Male | Normal | | details | | Urine | Detected and Quantified | 180.280 +/- 115.119 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 27.151 +/- 14.118 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 272.592 +/- 419.205 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 3.4 umol/mmol creatinine | Adult (>18 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 3397.0835 +/- 5584.336 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 616.862 +/- 982.852 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 7-120 umol/mmol creatinine | Children (1 - 13 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 7569.588 +/- 3599.0840 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 0.72-23.3 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 4.02(0.78-12.39) umol/mmol creatinine | Newborn (0-30 days old) | Female | Normal | | details | | Urine | Detected and Quantified | 3.14(0.71-23.84) umol/mmol creatinine | Newborn (0-30 days old) | Male | Normal | | details | | Urine | Detected and Quantified | 2.60 +/- 3.50 umol/mmol creatinine | Infant (0-1 year old) | Both | Normal | | details | | Urine | Detected and Quantified | 27.151 +/- 14.118 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 272.592 +/- 419.205 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 636.410 +/- 982.852 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 6.651 +/- 7.809 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 180.4 +/- 115.00 umol/mmol creatinine | Children (1-13 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 3397.0835 +/- 5584.336 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 7569.588 +/- 3599.0840 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 13.0 (1.0-27.0) umol/mmol creatinine | Children (1-13 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 1.4 +/- 1.3 umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 0.7 (0-2.0) umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | <21.17 umol/mmol creatinine | Children (1 - 18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 38.9 +/- 13.9 umol/mmol creatinine | Adult (>18 years old) | Male | Normal | | details | | Urine | Detected and Quantified | 32.3 +/- 8.7 umol/mmol creatinine | Adult (>18 years old) | Female | Normal | | details | | Urine | Detected and Quantified | 48.0 (0.00-200.0) umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 1.4-2.2 umol/mmol creatinine | Adult (>18 years old) | Male | Normal | | details | | Urine | Detected and Quantified | 1.62 (0.90-5.40) umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 3.6 (1.3-6.4) umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 2.257 +/- 1.254 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 2.0 (0.5-9.8) umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details | | Urine | Detected and Quantified | 1.4 (0.1-2.7) umol/mmol creatinine | Children (1-13 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 1.3 (0.1-5.8) umol/mmol creatinine | Adolescent (13-18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 29.712 +/- 14.532 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 0.62 (0.0-12.09) umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 2.24 (0.0-17.6) umol/mmol creatinine | Adult (>18 years old) | Female | Normal | | details | | Urine | Detected and Quantified | 3.6 (1.3-6.4) umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 2.0 (0.6-5.6) umol/mmol creatinine | Infant (0-1 year old) | Both | Normal | | details |
|
|---|
| Abnormal Concentrations |
|---|
| |
| Blood | Detected and Quantified | 235.0 +/- 17.0 uM | Adult (>18 years old) | Female | Obesity | | details | | Blood | Detected and Quantified | 35.1 +/- 33.9 uM | Adult (>18 years old) | Both | Heart Transplant | | details | | Blood | Detected and Quantified | 62.7 (85.0) uM | Adult (>18 years old) | Female | Down syndrome pregnancy | | details | | Blood | Detected and Quantified | 21.0 (19.4) uM | Adult (>18 years old) | Female | Pregnancy | | details | | Blood | Detected and Quantified | 500 uM | Adult (>18 years old) | Male | 3-Hydroxyacyl-CoA dehydrogenase deficiency (SCHAD) | | details | | Blood | Detected and Quantified | 660 uM | Adolescent (13-18 years old) | Female | 3-Hydroxyacyl-CoA dehydrogenase deficiency (SCHAD) | | details | | Blood | Detected and Quantified | 200-2800 uM | Infant (0-1 year old) | Female | 3-Hydroxyacyl-CoA dehydrogenase deficiency (SCHAD) | | details | | Blood | Detected and Quantified | 770-2900 uM | Children (1-13 years old) | Both | 3-Hydroxyacyl-CoA dehydrogenase deficiency (SCHAD) | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Oesophageal cancer | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Schizophrenia | | details | | Blood | Detected and Quantified | 220 uM | Infant (0-1 year old) | Not Specified | Very long-chain acyl-CoA dehydrogenase deficiency (vLCAD) | | details | | Blood | Detected and Quantified | 80-200 uM | Infant (0-1 year old) | Not Specified | Long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency | | details | | Blood | Detected and Quantified | 700-1340 uM | Infant (0-1 year old) | Not Specified | Medium chain acyl-CoA dehydrogenase deficiency (MCAD) | | details | | Blood | Detected and Quantified | 1800 uM | Infant (0-1 year old) | Male | Pyruvate dehydrogenase phosphatase deficiency | | details | | Blood | Detected and Quantified | <100 uM | Children (1-13 years old) | Male | Carnitine palmitoyltransferase deficiency I | | details | | Blood | Detected and Quantified | 1500.0 +/- 100.0 uM | Children (1-13 years old) | Both | Diabetes | | details | | Blood | Detected and Quantified | 0-380 uM | Infant (0-1 year old) | Not Specified | Short-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency | | details | | Blood | Detected and Quantified | 1490-5040 uM | Infant (0-1 year old) | Not Specified | Ketotic hypoglycemia | | details | | Blood | Detected and Quantified | 10-1380 uM | Infant (0-1 year old) | Female | 3-Hydroxyacyl-CoA dehydrogenase deficiency (SCHAD) | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Blood | Detected and Quantified | 35.5 +/- 92.2 uM | Adult (>18 years old) | Both | Diabetes | | details | | Blood | Detected and Quantified | 17.6 +/- 3.84 uM | Adult (>18 years old) | Both | Schizophrenia | | details | | Blood | Detected and Quantified | 7700.0 +/- 300.0 uM | Children (1-13 years old) | Both | Diabetes | | details | | Blood | Detected and Quantified | 54.57 +/- 64.53 uM | Adult (>18 years old) | Female | Pregnancy with fetuses with trisomy 18 | | details | | Blood | Detected and Quantified | 32.9 +/- 53.42 uM | Adult (>18 years old) | Female | Pregnancy | | details | | Blood | Detected and Quantified | 0-200 uM | Infant (0-1 year old) | Male | 3-Hydroxyacyl-CoA dehydrogenase deficiency (SCHAD) | | details | | Blood | Detected and Quantified | 31.6 (19.1) uM | Adult (>18 years old) | Female | Early preeclampsia | | details | | Blood | Detected and Quantified | 27.0 (16.2) uM | Adult (>18 years old) | Female | Pregnancy | | details | | Blood | Detected and Quantified | 64 uM | Infant (0-1 year old) | Female | 3-Hydroxy-3-Methylglutaryl-CoA Lyase Deficiency | | details | | Blood | Detected and Quantified | 100 uM | Newborn (0-30 days old) | Male | 2-Ketoglutarate dehydrogenase complex deficiency | | details | | Blood | Detected and Quantified | 120-2000 uM | Children (1-13 years old) | Male | 2-Ketoglutarate dehydrogenase complex deficiency | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Pancreatic cancer | | details | | Blood | Detected and Quantified | 33.63 (42.57) uM | Adult (>18 years old) | Female | Pregnancy with fetus having congenital heart defect | | details | | Blood | Detected and Quantified | 30.15 (37.69) uM | Adult (>18 years old) | Female | Pregnancy | | details | | Blood | Detected and Quantified | 29.61(44.27) uM | Adult (>18 years old) | Both | Heart failure with reduced ejection fraction | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Schizophrenia | | details | | Blood | Detected and Quantified | 49.9 (46.7) uM | Adult (>18 years old) | Female | Late-onset preeclampsia | | details | | Blood | Detected and Quantified | 29.7 (19.1) uM | Adult (>18 years old) | Female | Pregnancy | | details | | Blood | Detected and Quantified | 130 +/- 80 uM | Children (1-13 years old) | Both | Adrenal hyperplasia, congenital, due to 3-beta-hydroxysteroid dehydrogenase 2 deficiency | | details | | Blood | Detected and Quantified | 300 +/- 390 uM | Children (1-13 years old) | Both | Adrenal hyperplasia, congenital, due to 3-beta-hydroxysteroid dehydrogenase 2 deficiency | | details | | Blood | Detected and Quantified | 300 +/- 200 uM | Children (1-13 years old) | Both | Adrenal hyperplasia, congenital, due to 3-beta-hydroxysteroid dehydrogenase 2 deficiency | | details | | Blood | Detected and Quantified | 250 +/- 160 uM | Children (1-13 years old) | Both | Adrenal hyperplasia, congenital, due to 3-beta-hydroxysteroid dehydrogenase 2 deficiency | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | By NMR. Significantly decreased vs control. | | details | | Blood | Detected and Quantified | 1400.0 +/- 100.0 uM | Adult (>18 years old) | Both | Diabetes | | details | | Blood | Detected and Quantified | 7700.0 +/- 200.0 uM | Adult (>18 years old) | Both | Diabetes | | details | | Blood | Detected and Quantified | 5700 (4300-6000) uM | Adult (>18 years old) | Both | Diabetic ketoacidosis | | details | | Blood | Detected and Quantified | 20-50 uM | Children (1-13 years old) | Male | 3-Hydroxy-3-Methylglutaryl-CoA Synthase Deficiency | | details | | Cellular Cytoplasm | Detected and Quantified | 2600 (2500-2700) uM | Adult (>18 years old) | Both | Anoxia | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 430 (359-501) uM | Adult (>18 years old) | Both | Meningitis | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 6360 (5350-7370) uM | Adult (>18 years old) | Both | Diabetes | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal Cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Ulcerative colitis | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Crohn disease | | details | | Saliva | Detected and Quantified | 5.04 +/- 3.02 uM | Adult (>18 years old) | Both | Tooth alignment disorders | | details | | Saliva | Detected and Quantified | 6.62 +/- 0.72 uM | Adult (>18 years old) | Male | Alzheimer's disease | | details | | Saliva | Detected and Quantified | 6.37 +/- 1.75 uM | Adult (>18 years old) | Male | Frontotemporal lobe dementia | | details | | Saliva | Detected and Quantified | 24.25 +/- 32.08 uM | Adult (>18 years old) | Both | Lewy body disease | | details | | Urine | Detected and Quantified | 11.946 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Medium Chain Acyl-CoA Dehydrogenase Deficiency | | details | | Urine | Detected and Quantified | 16.29 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Medium Chain Acyl-CoA Dehydrogenase Deficiency | | details | | Urine | Detected and Quantified | 27.151 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Medium Chain Acyl-CoA Dehydrogenase Deficiency | | details | | Urine | Detected and Quantified | 31.495 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Medium Chain Acyl-CoA Dehydrogenase Deficiency | | details | | Urine | Detected and Quantified | 56.473 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Medium Chain Acyl-CoA Dehydrogenase Deficiency | | details | | Urine | Detected and Quantified | 351.872 umol/mmol creatinine | Infant (0-1 year old) | Male | Fumarase deficiency | | details | | Urine | Detected and Quantified | 45000 umol/mmol creatinine | Children (1-13 years old) | Not Available | Pyruvate carboxylase deficiency | | details | | Urine | Detected and Quantified | 8359 umol/mmol creatinine | Infant (0-1 year old) | Not Available | Pyruvate carboxylase deficiency | | details | | Urine | Detected and Quantified | 3-1050 umol/mmol creatinine | Children (1 - 13 years old) | Both | Medium Chain Acyl-CoA Dehydrogenase Deficiency | | details | | Urine | Detected and Quantified | 55.399-1634.282 umol/mmol creatinine | Children (1-13 years old) | Not Specified | Medium Chain Acyl-CoA Dehydrogenase Deficiency | | details | | Urine | Detected and Quantified | 170-390 umol/mmol creatinine | Children (1-13 years old) | Male | 3-Hydroxyisobutyric acid dehydrogenase deficiency | | details | | Urine | Detected and Quantified | 35.352 +/- 40.756 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Eosinophilic esophagitis | | details | | Urine | Detected and Quantified | 51.0431-56.473 umol/mmol creatinine | Infant (0-1 year old) | Not Specified | 3-Hydroxydicarboxylic aciduria | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Bladder cancer | | details | | Urine | Detected and Quantified | 224 umol/mmol creatinine | Adult (>18 years old) | Both | Diabetes | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Schizophrenia | | details | | Urine | Detected and Quantified | 30.883 +/- 22.266 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Eosinophilic esophagitis | | details | | Urine | Detected and Quantified | 74.311 +/- 155.898 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Gastroesophageal reflux disease | | details |
|
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | | Diabetes mellitus type 2 |
|---|
- Sheikh-Ali M, Karon BS, Basu A, Kudva YC, Muller LA, Xu J, Schwenk WF, Miles JM: Can serum beta-hydroxybutyrate be used to diagnose diabetic ketoacidosis? Diabetes Care. 2008 Apr;31(4):643-7. doi: 10.2337/dc07-1683. Epub 2008 Jan 9. [PubMed:18184896 ]
- Tasker RC, Lutman D, Peters MJ: Hyperventilation in severe diabetic ketoacidosis. Pediatr Crit Care Med. 2005 Jul;6(4):405-11. [PubMed:15982426 ]
- Bales JR, Higham DP, Howe I, Nicholson JK, Sadler PJ: Use of high-resolution proton nuclear magnetic resonance spectroscopy for rapid multi-component analysis of urine. Clin Chem. 1984 Mar;30(3):426-32. [PubMed:6321058 ]
- del Carmen Crespillo M, Olveira G, de Adana MS, Rojo-Martinez G, Garcia-Aleman J, Olvera P, Soriguer F, Munoz A: Metabolic effects of an enteral nutrition formula for diabetes: comparison with standard formulas in patients with type 1 diabetes. Clin Nutr. 2003 Oct;22(5):483-7. [PubMed:14512036 ]
| | Obesity |
|---|
- Vice E, Privette JD, Hickner RC, Barakat HA: Ketone body metabolism in lean and obese women. Metabolism. 2005 Nov;54(11):1542-5. [PubMed:16253646 ]
| | Pancreatic cancer |
|---|
- OuYang D, Xu J, Huang H, Chen Z: Metabolomic profiling of serum from human pancreatic cancer patients using 1H NMR spectroscopy and principal component analysis. Appl Biochem Biotechnol. 2011 Sep;165(1):148-54. doi: 10.1007/s12010-011-9240-0. Epub 2011 Apr 20. [PubMed:21505807 ]
- Zhang L, Jin H, Guo X, Yang Z, Zhao L, Tang S, Mo P, Wu K, Nie Y, Pan Y, Fan D: Distinguishing pancreatic cancer from chronic pancreatitis and healthy individuals by (1)H nuclear magnetic resonance-based metabonomic profiles. Clin Biochem. 2012 Sep;45(13-14):1064-9. doi: 10.1016/j.clinbiochem.2012.05.012. Epub 2012 May 19. [PubMed:22613268 ]
| | Colorectal cancer |
|---|
- Ni Y, Xie G, Jia W: Metabonomics of human colorectal cancer: new approaches for early diagnosis and biomarker discovery. J Proteome Res. 2014 Sep 5;13(9):3857-70. doi: 10.1021/pr500443c. Epub 2014 Aug 14. [PubMed:25105552 ]
- Brown DG, Rao S, Weir TL, O'Malia J, Bazan M, Brown RJ, Ryan EP: Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016 Jun 6;4:11. doi: 10.1186/s40170-016-0151-y. eCollection 2016. [PubMed:27275383 ]
| | Schizophrenia |
|---|
- Fukushima T, Iizuka H, Yokota A, Suzuki T, Ohno C, Kono Y, Nishikiori M, Seki A, Ichiba H, Watanabe Y, Hongo S, Utsunomiya M, Nakatani M, Sadamoto K, Yoshio T: Quantitative analyses of schizophrenia-associated metabolites in serum: serum D-lactate levels are negatively correlated with gamma-glutamylcysteine in medicated schizophrenia patients. PLoS One. 2014 Jul 8;9(7):e101652. doi: 10.1371/journal.pone.0101652. eCollection 2014. [PubMed:25004141 ]
- Yang J, Chen T, Sun L, Zhao Z, Qi X, Zhou K, Cao Y, Wang X, Qiu Y, Su M, Zhao A, Wang P, Yang P, Wu J, Feng G, He L, Jia W, Wan C: Potential metabolite markers of schizophrenia. Mol Psychiatry. 2013 Jan;18(1):67-78. doi: 10.1038/mp.2011.131. Epub 2011 Oct 25. [PubMed:22024767 ]
- Cai HL, Li HD, Yan XZ, Sun B, Zhang Q, Yan M, Zhang WY, Jiang P, Zhu RH, Liu YP, Fang PF, Xu P, Yuan HY, Zhang XH, Hu L, Yang W, Ye HS: Metabolomic analysis of biochemical changes in the plasma and urine of first-episode neuroleptic-naive schizophrenia patients after treatment with risperidone. J Proteome Res. 2012 Aug 3;11(8):4338-50. doi: 10.1021/pr300459d. Epub 2012 Jul 26. [PubMed:22800120 ]
| | Early preeclampsia |
|---|
- Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K: Metabolomics and first-trimester prediction of early-onset preeclampsia. J Matern Fetal Neonatal Med. 2012 Oct;25(10):1840-7. doi: 10.3109/14767058.2012.680254. Epub 2012 Apr 28. [PubMed:22494326 ]
| | Pregnancy |
|---|
- Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K: Metabolomics and first-trimester prediction of early-onset preeclampsia. J Matern Fetal Neonatal Med. 2012 Oct;25(10):1840-7. doi: 10.3109/14767058.2012.680254. Epub 2012 Apr 28. [PubMed:22494326 ]
- Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K: First-trimester metabolomic detection of late-onset preeclampsia. Am J Obstet Gynecol. 2013 Jan;208(1):58.e1-7. doi: 10.1016/j.ajog.2012.11.003. Epub 2012 Nov 13. [PubMed:23159745 ]
- Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K: Metabolomic analysis for first-trimester Down syndrome prediction. Am J Obstet Gynecol. 2013 May;208(5):371.e1-8. doi: 10.1016/j.ajog.2012.12.035. Epub 2013 Jan 8. [PubMed:23313728 ]
- Bahado-Singh RO, Akolekar R, Chelliah A, Mandal R, Dong E, Kruger M, Wishart DS, Nicolaides K: Metabolomic analysis for first-trimester trisomy 18 detection. Am J Obstet Gynecol. 2013 Jul;209(1):65.e1-9. doi: 10.1016/j.ajog.2013.03.028. Epub 2013 Mar 25. [PubMed:23535240 ]
- Bahado-Singh RO, Ertl R, Mandal R, Bjorndahl TC, Syngelaki A, Han B, Dong E, Liu PB, Alpay-Savasan Z, Wishart DS, Nicolaides KH: Metabolomic prediction of fetal congenital heart defect in the first trimester. Am J Obstet Gynecol. 2014 Sep;211(3):240.e1-240.e14. doi: 10.1016/j.ajog.2014.03.056. Epub 2014 Apr 1. [PubMed:24704061 ]
| | Late-onset preeclampsia |
|---|
- Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K: First-trimester metabolomic detection of late-onset preeclampsia. Am J Obstet Gynecol. 2013 Jan;208(1):58.e1-7. doi: 10.1016/j.ajog.2012.11.003. Epub 2012 Nov 13. [PubMed:23159745 ]
| | 2-Ketoglutarate dehydrogenase complex deficiency |
|---|
- Guffon N, Lopez-Mediavilla C, Dumoulin R, Mousson B, Godinot C, Carrier H, Collombet JM, Divry P, Mathieu M, Guibaud P: 2-Ketoglutarate dehydrogenase deficiency, a rare cause of primary hyperlactataemia: report of a new case. J Inherit Metab Dis. 1993;16(5):821-30. [PubMed:8295396 ]
| | 3-Hydroxy-3-Methylglutaryl-CoA Synthase Deficiency |
|---|
- Morris AA, Lascelles CV, Olpin SE, Lake BD, Leonard JV, Quant PA: Hepatic mitochondrial 3-hydroxy-3-methylglutaryl-coenzyme a synthase deficiency. Pediatr Res. 1998 Sep;44(3):392-6. doi: 10.1203/00006450-199809000-00021. [PubMed:9727719 ]
| | 3-Hydroxyacyl-CoA dehydrogenase deficiency |
|---|
- Hsu BY, Kelly A, Thornton PS, Greenberg CR, Dilling LA, Stanley CA: Protein-sensitive and fasting hypoglycemia in children with the hyperinsulinism/hyperammonemia syndrome. J Pediatr. 2001 Mar;138(3):383-9. doi: 10.1067/mpd.2001.111818. [PubMed:11241047 ]
- Clayton PT, Eaton S, Aynsley-Green A, Edginton M, Hussain K, Krywawych S, Datta V, Malingre HE, Berger R, van den Berg IE: Hyperinsulinism in short-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency reveals the importance of beta-oxidation in insulin secretion. J Clin Invest. 2001 Aug;108(3):457-65. [PubMed:11489939 ]
- Popa FI, Perlini S, Teofoli F, Degani D, Funghini S, La Marca G, Rinaldo P, Vincenzi M, Antoniazzi F, Boner A, Camilot M: 3-hydroxyacyl-coenzyme a dehydrogenase deficiency: identification of a new mutation causing hyperinsulinemic hypoketotic hypoglycemia, altered organic acids and acylcarnitines concentrations. JIMD Rep. 2012;2:71-7. doi: 10.1007/8904_2011_50. Epub 2011 Sep 6. [PubMed:23430856 ]
| | Long-chain-3-hydroxyacyl CoA dehydrogenase deficiency |
|---|
- Clayton PT, Eaton S, Aynsley-Green A, Edginton M, Hussain K, Krywawych S, Datta V, Malingre HE, Berger R, van den Berg IE: Hyperinsulinism in short-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency reveals the importance of beta-oxidation in insulin secretion. J Clin Invest. 2001 Aug;108(3):457-65. [PubMed:11489939 ]
| | Short-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency |
|---|
- Clayton PT, Eaton S, Aynsley-Green A, Edginton M, Hussain K, Krywawych S, Datta V, Malingre HE, Berger R, van den Berg IE: Hyperinsulinism in short-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency reveals the importance of beta-oxidation in insulin secretion. J Clin Invest. 2001 Aug;108(3):457-65. [PubMed:11489939 ]
| | Ketotic hypoglycemia |
|---|
- Clayton PT, Eaton S, Aynsley-Green A, Edginton M, Hussain K, Krywawych S, Datta V, Malingre HE, Berger R, van den Berg IE: Hyperinsulinism in short-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency reveals the importance of beta-oxidation in insulin secretion. J Clin Invest. 2001 Aug;108(3):457-65. [PubMed:11489939 ]
| | 3-Hydroxy-3-methylglutaryl-CoA lyase deficiency |
|---|
- Zschocke J, Penzien JM, Bielen R, Casals N, Aledo R, Pie J, Hoffmann GF, Hegardt FG, Mayatepek E: The diagnosis of mitochondrial HMG-CoA synthase deficiency. J Pediatr. 2002 Jun;140(6):778-80. doi: 10.1067/mpd.2002.123854. [PubMed:12072887 ]
| | Adrenal hyperplasia, congenital, due to 3-beta-hydroxysteroid dehydrogenase 2 deficiency |
|---|
- Benkert AR, Young M, Robinson D, Hendrickson C, Lee PA, Strauss KA: Severe Salt-Losing 3beta-Hydroxysteroid Dehydrogenase Deficiency: Treatment and Outcomes of HSD3B2 c.35G>A Homozygotes. J Clin Endocrinol Metab. 2015 Aug;100(8):E1105-15. doi: 10.1210/jc.2015-2098. Epub 2015 Jun 16. [PubMed:26079780 ]
| | Pyruvate dehydrogenase phosphatase deficiency |
|---|
- Robinson BH, Sherwood WG: Pyruvate dehydrogenase phosphatase deficiency: a cause of congenital chronic lactic acidosis in infancy. Pediatr Res. 1975 Dec;9(12):935-9. doi: 10.1203/00006450-197512000-00015. [PubMed:172850 ]
| | Carnitine palmitoyltransferase I deficiency |
|---|
- Olpin SE, Allen J, Bonham JR, Clark S, Clayton PT, Calvin J, Downing M, Ives K, Jones S, Manning NJ, Pollitt RJ, Standing SJ, Tanner MS: Features of carnitine palmitoyltransferase type I deficiency. J Inherit Metab Dis. 2001 Feb;24(1):35-42. [PubMed:11286380 ]
| | Anoxia |
|---|
- Zupke C, Sinskey AJ, Stephanopoulos G: Intracellular flux analysis applied to the effect of dissolved oxygen on hybridomas. Appl Microbiol Biotechnol. 1995 Dec;44(1-2):27-36. [PubMed:8579834 ]
| | Meningitis |
|---|
- Subramanian A, Gupta A, Saxena S, Gupta A, Kumar R, Nigam A, Kumar R, Mandal SK, Roy R: Proton MR CSF analysis and a new software as predictors for the differentiation of meningitis in children. NMR Biomed. 2005 Jun;18(4):213-25. [PubMed:15627241 ]
| | Ulcerative colitis |
|---|
- Azario I, Pievani A, Del Priore F, Antolini L, Santi L, Corsi A, Cardinale L, Sawamoto K, Kubaski F, Gentner B, Bernardo ME, Valsecchi MG, Riminucci M, Tomatsu S, Aiuti A, Biondi A, Serafini M: Neonatal umbilical cord blood transplantation halts skeletal disease progression in the murine model of MPS-I. Sci Rep. 2017 Aug 25;7(1):9473. doi: 10.1038/s41598-017-09958-9. [PubMed:28842642 ]
| | Crohn's disease |
|---|
- Azario I, Pievani A, Del Priore F, Antolini L, Santi L, Corsi A, Cardinale L, Sawamoto K, Kubaski F, Gentner B, Bernardo ME, Valsecchi MG, Riminucci M, Tomatsu S, Aiuti A, Biondi A, Serafini M: Neonatal umbilical cord blood transplantation halts skeletal disease progression in the murine model of MPS-I. Sci Rep. 2017 Aug 25;7(1):9473. doi: 10.1038/s41598-017-09958-9. [PubMed:28842642 ]
| | Alzheimer's disease |
|---|
- Tsuruoka M, Hara J, Hirayama A, Sugimoto M, Soga T, Shankle WR, Tomita M: Capillary electrophoresis-mass spectrometry-based metabolome analysis of serum and saliva from neurodegenerative dementia patients. Electrophoresis. 2013 Oct;34(19):2865-72. doi: 10.1002/elps.201300019. Epub 2013 Sep 6. [PubMed:23857558 ]
| | Frontotemporal dementia |
|---|
- Tsuruoka M, Hara J, Hirayama A, Sugimoto M, Soga T, Shankle WR, Tomita M: Capillary electrophoresis-mass spectrometry-based metabolome analysis of serum and saliva from neurodegenerative dementia patients. Electrophoresis. 2013 Oct;34(19):2865-72. doi: 10.1002/elps.201300019. Epub 2013 Sep 6. [PubMed:23857558 ]
| | Lewy body disease |
|---|
- Tsuruoka M, Hara J, Hirayama A, Sugimoto M, Soga T, Shankle WR, Tomita M: Capillary electrophoresis-mass spectrometry-based metabolome analysis of serum and saliva from neurodegenerative dementia patients. Electrophoresis. 2013 Oct;34(19):2865-72. doi: 10.1002/elps.201300019. Epub 2013 Sep 6. [PubMed:23857558 ]
| | Eosinophilic esophagitis |
|---|
- Slae, M., Huynh, H., Wishart, D.S. (2014). Analysis of 30 normal pediatric urine samples via NMR spectroscopy (unpublished work). NA.
| | Medium Chain Acyl-CoA Dehydrogenase Deficiency |
|---|
- Tserng KY, Jin SJ, Kerr DS, Hoppel CL: Abnormal urinary excretion of unsaturated dicarboxylic acids in patients with medium-chain acyl-CoA dehydrogenase deficiency. J Lipid Res. 1990 May;31(5):763-71. [PubMed:2380628 ]
- Gregersen N, Kolvraa S, Rasmussen K, Mortensen PB, Divry P, David M, Hobolth N: General (medium-chain) acyl-CoA dehydrogenase deficiency (non-ketotic dicarboxylic aciduria): quantitative urinary excretion pattern of 23 biologically significant organic acids in three cases. Clin Chim Acta. 1983 Aug 15;132(2):181-91. [PubMed:6616873 ]
- Duran M, De Klerk JB, Wadman SK, Bruinvis L, Ketting D: The differential diagnosis of dicarboxylic aciduria. J Inherit Metab Dis. 1984;7 Suppl 1:48-51. [PubMed:6434845 ]
- Clayton PT, Eaton S, Aynsley-Green A, Edginton M, Hussain K, Krywawych S, Datta V, Malingre HE, Berger R, van den Berg IE: Hyperinsulinism in short-chain L-3-hydroxyacyl-CoA dehydrogenase deficiency reveals the importance of beta-oxidation in insulin secretion. J Clin Invest. 2001 Aug;108(3):457-65. [PubMed:11489939 ]
| | Fumarase deficiency |
|---|
- Bastug O, Kardas F, Ozturk MA, Halis H, Memur S, Korkmaz L, Tag Z, Gunes T: A rare cause of opistotonus; fumaric aciduria: The first case presentation in Turkey. Turk Pediatri Ars. 2014 Mar 1;49(1):74-6. doi: 10.5152/tpa.2014.442. eCollection 2014 Mar. [PubMed:26078636 ]
| | 3-Hydroxyisobutyric acid dehydrogenase deficiency |
|---|
- Ko FJ, Nyhan WL, Wolff J, Barshop B, Sweetman L: 3-Hydroxyisobutyric aciduria: an inborn error of valine metabolism. Pediatr Res. 1991 Oct;30(4):322-6. doi: 10.1203/00006450-199110000-00006. [PubMed:1956714 ]
| | 3-Hydroxydicarboxylic aciduria |
|---|
- Tserng KY, Jin SJ, Kerr DS, Hoppel CL: Urinary 3-hydroxydicarboxylic acids in pathophysiology of metabolic disorders with dicarboxylic aciduria. Metabolism. 1991 Jul;40(7):676-82. [PubMed:1870421 ]
| | Pyruvate carboxylase deficiency |
|---|
- Habarou F, Brassier A, Rio M, Chretien D, Monnot S, Barbier V, Barouki R, Bonnefont JP, Boddaert N, Chadefaux-Vekemans B, Le Moyec L, Bastin J, Ottolenghi C, de Lonlay P: Pyruvate carboxylase deficiency: An underestimated cause of lactic acidosis. Mol Genet Metab Rep. 2014 Nov 28;2:25-31. doi: 10.1016/j.ymgmr.2014.11.001. eCollection 2015 Mar. [PubMed:28649521 ]
|
|
|---|
| Associated OMIM IDs | - 125853 (Diabetes mellitus type 2)
- 601665 (Obesity)
- 260350 (Pancreatic cancer)
- 114500 (Colorectal cancer)
- 181500 (Schizophrenia)
- 203740 (2-Ketoglutarate dehydrogenase complex deficiency)
- 605911 (3-Hydroxy-3-Methylglutaryl-CoA Synthase Deficiency)
- 231530 (3-Hydroxyacyl-CoA dehydrogenase deficiency)
- 246450 (3-Hydroxy-3-methylglutaryl-CoA lyase deficiency)
- 201810 (Adrenal hyperplasia, congenital, due to 3-beta-hydroxysteroid dehydrogenase 2 deficiency)
- 608782 (Pyruvate dehydrogenase phosphatase deficiency)
- 255120 (Carnitine palmitoyltransferase I deficiency)
- 266600 (Crohn's disease)
- 104300 (Alzheimer's disease)
- 600274 (Frontotemporal dementia)
- 610247 (Eosinophilic esophagitis)
- 201450 (Medium Chain Acyl-CoA Dehydrogenase Deficiency)
- 606812 (Fumarase deficiency)
- 266150 (Pyruvate carboxylase deficiency)
|
|---|
| External Links |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB021869 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 83181 |
|---|
| KEGG Compound ID | C01089 |
|---|
| BioCyc ID | CPD-335 |
|---|
| BiGG ID | 36784 |
|---|
| Wikipedia Link | Beta-Hydroxybutyric_acid |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 92135 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17066 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | BHB |
|---|
| MarkerDB ID | MDB00000005 |
|---|
| Good Scents ID | Not Available |
|---|
| References |
|---|
| Synthesis Reference | Le Sann, Christine; Munoz, Dulce M.; Saunders, Natalie; Simpson, Thomas J.; Smith, David I.; Soulas, Florilene; Watts, Paul; Willis, Christine L. Assembly intermediates in polyketide biosynthesis: enantioselective syntheses of b-hydroxycarbonyl compounds. Org Biomol Chem. 2005 May 7;3(9):1719-28. Epub 2005 Mar 31. Pubmed: 15858656 |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| General References | - Pan JW, Rothman TL, Behar KL, Stein DT, Hetherington HP: Human brain beta-hydroxybutyrate and lactate increase in fasting-induced ketosis. J Cereb Blood Flow Metab. 2000 Oct;20(10):1502-7. [PubMed:11043913 ]
- Pan JW, Telang FW, Lee JH, de Graaf RA, Rothman DL, Stein DT, Hetherington HP: Measurement of beta-hydroxybutyrate in acute hyperketonemia in human brain. J Neurochem. 2001 Nov;79(3):539-44. [PubMed:11701757 ]
- Plecko B, Stoeckler-Ipsiroglu S, Schober E, Harrer G, Mlynarik V, Gruber S, Moser E, Moeslinger D, Silgoner H, Ipsiroglu O: Oral beta-hydroxybutyrate supplementation in two patients with hyperinsulinemic hypoglycemia: monitoring of beta-hydroxybutyrate levels in blood and cerebrospinal fluid, and in the brain by in vivo magnetic resonance spectroscopy. Pediatr Res. 2002 Aug;52(2):301-6. [PubMed:12149510 ]
- Raunio RP, Leivo PV, Kuusinen AM: Bioluminescent assay of D-3-hydroxybutyrate in serum. J Biolumin Chemilumin. 1986 Jun;1(1):11-4. [PubMed:3503522 ]
- Shoemaker JD, Elliott WH: Automated screening of urine samples for carbohydrates, organic and amino acids after treatment with urease. J Chromatogr. 1991 Jan 2;562(1-2):125-38. [PubMed:2026685 ]
- Hoffmann GF, Meier-Augenstein W, Stockler S, Surtees R, Rating D, Nyhan WL: Physiology and pathophysiology of organic acids in cerebrospinal fluid. J Inherit Metab Dis. 1993;16(4):648-69. [PubMed:8412012 ]
- Zupke C, Sinskey AJ, Stephanopoulos G: Intracellular flux analysis applied to the effect of dissolved oxygen on hybridomas. Appl Microbiol Biotechnol. 1995 Dec;44(1-2):27-36. [PubMed:8579834 ]
- Redjems-Bennani N, Jeandel C, Lefebvre E, Blain H, Vidailhet M, Gueant JL: Abnormal substrate levels that depend upon mitochondrial function in cerebrospinal fluid from Alzheimer patients. Gerontology. 1998;44(5):300-4. [PubMed:9693263 ]
- Frenkel G, Peterson RN, Freund M: Oxidative and glycolytic metabolism of semen components by washed guinea pig spermatozoa. Fertil Steril. 1975 Feb;26(2):144-7. [PubMed:1126459 ]
- Abrahamsson K, Eriksson BO, Holme E, Jodal U, Jonsson A, Lindstedt S: Pivalic acid-induced carnitine deficiency and physical exercise in humans. Metabolism. 1996 Dec;45(12):1501-7. [PubMed:8969283 ]
- Teresinski G, Buszewicz G, Madro R: The influence of ethanol on the level of ketone bodies in hypothermia. Forensic Sci Int. 2002 Jun 25;127(1-2):88-96. [PubMed:12098531 ]
- Nalecz KA, Miecz D, Berezowski V, Cecchelli R: Carnitine: transport and physiological functions in the brain. Mol Aspects Med. 2004 Oct-Dec;25(5-6):551-67. [PubMed:15363641 ]
- Blomqvist G, Alvarsson M, Grill V, Von Heijne G, Ingvar M, Thorell JO, Stone-Elander S, Widen L, Ekberg K: Effect of acute hyperketonemia on the cerebral uptake of ketone bodies in nondiabetic subjects and IDDM patients. Am J Physiol Endocrinol Metab. 2002 Jul;283(1):E20-8. [PubMed:12067838 ]
- Krotkiewski M: Value of VLCD supplementation with medium chain triglycerides. Int J Obes Relat Metab Disord. 2001 Sep;25(9):1393-400. [PubMed:11571605 ]
- Kamijo T, Indo Y, Souri M, Aoyama T, Hara T, Yamamoto S, Ushikubo S, Rinaldo P, Matsuda I, Komiyama A, Hashimoto T: Medium chain 3-ketoacyl-coenzyme A thiolase deficiency: a new disorder of mitochondrial fatty acid beta-oxidation. Pediatr Res. 1997 Nov;42(5):569-76. [PubMed:9357925 ]
- Geary N, Grotschel H, Scharrer E: Blood metabolites and feeding during postinsulin hypophagia. Am J Physiol. 1982 Sep;243(3):R304-11. [PubMed:7051864 ]
- Yeh YY, Sheehan PM: Preferential utilization of ketone bodies in the brain and lung of newborn rats. Fed Proc. 1985 Apr;44(7):2352-8. [PubMed:3884391 ]
- Kita K, Ishimaru K, Teraoka M, Yanase H, Kato N: Properties of poly(3-hydroxybutyrate) depolymerase from a marine bacterium, Alcaligenes faecalis AE122. Appl Environ Microbiol. 1995 May;61(5):1727-30. [PubMed:7646009 ]
- Kim BS, Lee SC, Lee SY, Chang HN, Chang YK, Woo SI: Production of poly(3-hydroxybutyric acid) by fed-batch culture of Alcaligenes eutrophus with glucose concentration control. Biotechnol Bioeng. 1994 Apr 15;43(9):892-8. doi: 10.1002/bit.260430908. [PubMed:18615882 ]
- Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. [PubMed:32033212 ]
- (). Bernardi, R. et al., Chem. Commun., 1984, 460, (resoln). .
- (). Encyclopedia of Food and Color Additives, (ed. Burdock, G.A.), CRC Press, 1997, 994-995, (Et ester). .
- (). Fenaroli's Handbook of Flavor Ingredients, 3rd edn., (ed. Burdock, G.A.), CRC Press, 1995, 1, 238, (Et ester). .
- (). Forsyth, W.G.C. et al., Nature (London), 1958, 182, 800, (isol). .
- (). Gottschalk, G. et al., Nature (London), 1965, 205, 308. .
- (). Krajewski, D. et al., Phytochemistry, 1997, 45, 1627, (ester glucosides). .
- (). Levene, P.A. et al., J. Biol. Chem., 1926, 68, 418, (abs config). .
- (). Lewis, R.J., Sax's Dangerous Properties of Industrial Materials, 8th edn., Van Nostrand Reinhold, 1992, MEF500 MKP250. .
- (). Marchessault, R.H. et al., Makromol. Chem., Macromol. Symp., 1988, 19, 235, (rev, polymers). .
- (). Muller, H.-M. et al., Angew. Chem., Int. Ed., 1993, 32, 477, (rev, polymer). .
- (). Org. Synth., 1993, 71, 39, (synth, acid, Meester, pmr, ir). .
- (). Quang DN, Hashimoto T, Toyota M, Asakawa Y. Occurrence of a high concentration of spider pheromones in the ascomycete fungus Hypoxylon truncatum. J Nat Prod. 2003 Dec;66(12):1613-4.. .
- (). Schulz S, Toft S. Identification of a sex pheromone from a spider. Science. 1993 Jun 11;260(5114):1635-7.. .
- (). Seebach, D. et al., Helv. Chim. Acta, 1982, 65, 495-503, (esters, synth). .
- (). Seebach, D. et al., Helv. Chim. Acta, 1988, 71, 155, (synth, dimer). .
- (). Wipf, B. et al., Helv. Chim. Acta, 1983, 66, 485. .
|
|---|