| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:49:05 UTC |
|---|
| HMDB ID | HMDB0000698 |
|---|
| Secondary Accession Numbers | - HMDB0002474
- HMDB00698
- HMDB02474
|
|---|
| Metabolite Identification |
|---|
| Common Name | Lithocholic acid glycine conjugate |
|---|
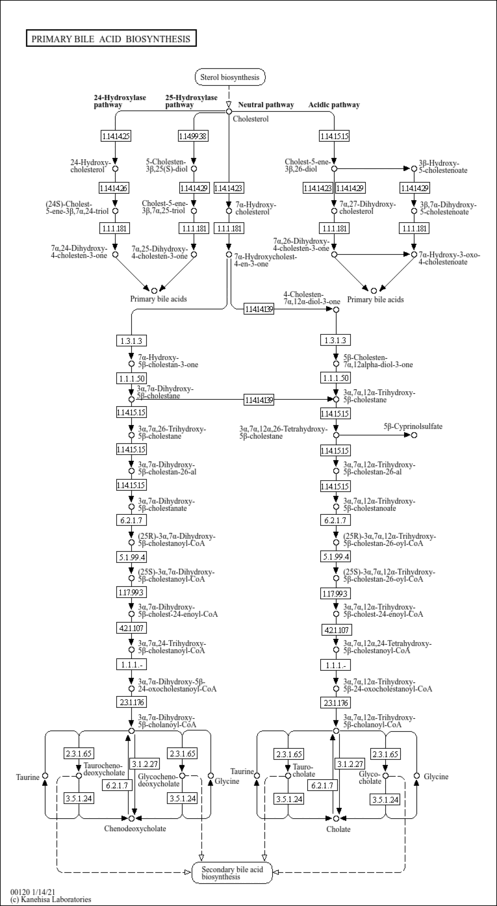
| Description | Lithocholic acid glycine conjugate is an acyl glycine and a bile acid-glycine conjugate. is an acyl glycine and a bile acid-glycine conjugate. It is a secondary bile acid produced by the action of enzymes existing in the microbial flora of the colonic environment. In hepatocytes, both primary and secondary bile acids undergo amino acid conjugation at the C-24 carboxylic acid on the side chain, and almost all bile acids in the bile duct therefore exist in a glycine conjugated form (PMID: 16949895 ). Bile acids are steroid acids found predominantly in the bile of mammals. The distinction between different bile acids is minute, depending only on the presence or absence of hydroxyl groups on positions 3, 7, and 12. Bile acids are physiological detergents that facilitate excretion, absorption, and transport of fats and sterols in the intestine and liver. Bile acids are also steroidal amphipathic molecules derived from the catabolism of cholesterol. They modulate bile flow and lipid secretion, are essential for the absorption of dietary fats and vitamins, and have been implicated in the regulation of all the key enzymes involved in cholesterol homeostasis. Bile acids recirculate through the liver, bile ducts, small intestine and portal vein to form an enterohepatic circuit. They exist as anions at physiological pH and, consequently, require a carrier for transport across the membranes of the enterohepatic tissues. The unique detergent properties of bile acids are essential for the digestion and intestinal absorption of hydrophobic nutrients. Bile acids have potent toxic properties (e.g. membrane disruption) and there are a plethora of mechanisms to limit their accumulation in blood and tissues (PMID: 11316487 , 16037564 , 12576301 , 11907135 ). |
|---|
| Structure | [H][C@@]1(CC[C@@]2([H])[C@]3([H])CC[C@]4([H])C[C@H](O)CC[C@]4(C)[C@@]3([H])CC[C@]12C)[C@H](C)CCC(=O)NCC(O)=O InChI=1S/C26H43NO4/c1-16(4-9-23(29)27-15-24(30)31)20-7-8-21-19-6-5-17-14-18(28)10-12-25(17,2)22(19)11-13-26(20,21)3/h16-22,28H,4-15H2,1-3H3,(H,27,29)(H,30,31)/t16-,17-,18-,19+,20-,21+,22+,25+,26-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| Glycolithocholate | ChEBI | | Lithocholylglycine | ChEBI | | N-((3-alpha,5-beta)-3-Hydroxy-24-oxocholan-24-yl)glycine | ChEBI | | Glycolithocholic acid | Kegg | | N-((3-a,5-b)-3-Hydroxy-24-oxocholan-24-yl)glycine | Generator | | N-((3-Α,5-β)-3-hydroxy-24-oxocholan-24-yl)glycine | Generator | | Lithocholate glycine conjugate | Generator | | Lithocholic acid glycine conjugic acid | Generator | | 3a-Hydroxy-5b-cholanic acid glycine ester | HMDB | | 3a-Hydroxy-5b-cholanoylglycine | HMDB | | 3a-Hydroxy-N-(carboxymethyl)-5b-cholan-24-amide | HMDB | | 3alpha-Hydroxy-5beta-cholan-24-Oate | HMDB | | 3alpha-Hydroxy-5beta-cholan-24-Oic acid | HMDB | | 3alpha-Hydroxy-5beta-cholan-24-Oic acid N-(carboxymethyl)amide | HMDB | | Lithocholoyglycine | HMDB | | N-(3a-Hydroxy-5b-cholan-24-oyl)-glycine | HMDB | | N-(3a-Hydroxy-5b-cholanoyl)glycine | HMDB | | N-(Carboxymethyl)-3a-hydroxy-5b-cholan-24-amide | HMDB | | Glycolithocholic acid, monosodium salt | HMDB | | 3alpha-Hydroxy-5beta-cholanoylglycine 3-sulfate | HMDB | | 3alpha-Hydroxy-5beta-cholanoylglycine 3-sulphate | HMDB | | 3α-Hydroxy-5β-cholanoylglycine 3-sulfate | HMDB | | 3α-Hydroxy-5β-cholanoylglycine 3-sulphate | HMDB | | Glycolithocholic acid 3-sulfate | HMDB | | Glycolithocholic acid 3-sulphate | HMDB | | Glycolithocholic acid 3alpha-sulfate | HMDB | | Glycolithocholic acid 3alpha-sulphate | HMDB | | Glycolithocholic acid 3α-sulfate | HMDB | | Glycolithocholic acid 3α-sulphate | HMDB | | Glycolithocholic acid sulfate | HMDB | | Glycolithocholic acid sulphate | HMDB | | N-[(3alpha,5beta)-24-Oxo-3-(sulfooxy)cholan-24-yl]glycine | HMDB | | N-[(3α,5β)-24-Oxo-3-(sulfooxy)cholan-24-yl]glycine | HMDB | | Sulfoglycolithocholic acid | HMDB | | Sulfolithocholylglycine | HMDB |
|
|---|
| Chemical Formula | C26H43NO4 |
|---|
| Average Molecular Weight | 433.6239 |
|---|
| Monoisotopic Molecular Weight | 433.319208869 |
|---|
| IUPAC Name | 2-[(4R)-4-[(1S,2S,5R,7R,10R,11S,14R,15R)-5-hydroxy-2,15-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanamido]acetic acid |
|---|
| Traditional Name | [(4R)-4-[(1S,2S,5R,7R,10R,11S,14R,15R)-5-hydroxy-2,15-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanamido]acetic acid |
|---|
| CAS Registry Number | 474-74-8 |
|---|
| SMILES | [H][C@@]1(CC[C@@]2([H])[C@]3([H])CC[C@]4([H])C[C@H](O)CC[C@]4(C)[C@@]3([H])CC[C@]12C)[C@H](C)CCC(=O)NCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C26H43NO4/c1-16(4-9-23(29)27-15-24(30)31)20-7-8-21-19-6-5-17-14-18(28)10-12-25(17,2)22(19)11-13-26(20,21)3/h16-22,28H,4-15H2,1-3H3,(H,27,29)(H,30,31)/t16-,17-,18-,19+,20-,21+,22+,25+,26-/m1/s1 |
|---|
| InChI Key | XBSQTYHEGZTYJE-OETIFKLTSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as glycinated bile acids and derivatives. Glycinated bile acids and derivatives are compounds with a structure characterized by the presence of a glycine linked to a bile acid skeleton. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Glycinated bile acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycinated bile acid
- Hydroxy bile acid, alcohol, or derivatives
- Monohydroxy bile acid, alcohol, or derivatives
- 3-hydroxysteroid
- Hydroxysteroid
- 3-alpha-hydroxysteroid
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- Alpha-amino acid or derivatives
- Cyclic alcohol
- Secondary alcohol
- Carboximidic acid
- Carboximidic acid derivative
- Carboxylic acid derivative
- Carboxylic acid
- Organic 1,3-dipolar compound
- Monocarboxylic acid or derivatives
- Propargyl-type 1,3-dipolar organic compound
- Organopnictogen compound
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Carbonyl group
- Organic nitrogen compound
- Alcohol
- Organooxygen compound
- Organonitrogen compound
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 7.91 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 16.8866 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 2.37 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 72.8 seconds | 40023050 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 2927.2 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 265.6 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 233.1 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 178.3 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 630.5 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 709.5 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 720.0 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 128.9 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 1284.7 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 584.9 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 1758.1 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 416.1 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 495.6 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 229.4 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 268.7 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 21.0 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Lithocholic acid glycine conjugate,1TMS,isomer #1 | C[C@H](CCC(=O)NCC(=O)O)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 3838.0 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,1TMS,isomer #2 | C[C@H](CCC(=O)NCC(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 3776.5 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,1TMS,isomer #3 | C[C@H](CCC(=O)N(CC(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 3830.9 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,2TMS,isomer #1 | C[C@H](CCC(=O)NCC(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 3759.0 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,2TMS,isomer #2 | C[C@H](CCC(=O)N(CC(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 3803.9 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,2TMS,isomer #3 | C[C@H](CCC(=O)N(CC(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 3753.7 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,3TMS,isomer #1 | C[C@H](CCC(=O)N(CC(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 3724.0 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,3TMS,isomer #1 | C[C@H](CCC(=O)N(CC(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 3763.7 | Standard non polar | 33892256 | | Lithocholic acid glycine conjugate,3TMS,isomer #1 | C[C@H](CCC(=O)N(CC(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4223.5 | Standard polar | 33892256 | | Lithocholic acid glycine conjugate,1TBDMS,isomer #1 | C[C@H](CCC(=O)NCC(=O)O)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4065.3 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,1TBDMS,isomer #2 | C[C@H](CCC(=O)NCC(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4052.6 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,1TBDMS,isomer #3 | C[C@H](CCC(=O)N(CC(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4058.5 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,2TBDMS,isomer #1 | C[C@H](CCC(=O)NCC(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4253.5 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,2TBDMS,isomer #2 | C[C@H](CCC(=O)N(CC(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4264.7 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,2TBDMS,isomer #3 | C[C@H](CCC(=O)N(CC(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4245.0 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,3TBDMS,isomer #1 | C[C@H](CCC(=O)N(CC(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4435.9 | Semi standard non polar | 33892256 | | Lithocholic acid glycine conjugate,3TBDMS,isomer #1 | C[C@H](CCC(=O)N(CC(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4404.7 | Standard non polar | 33892256 | | Lithocholic acid glycine conjugate,3TBDMS,isomer #1 | C[C@H](CCC(=O)N(CC(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4432.2 | Standard polar | 33892256 |
|
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (Non-derivatized) - 70eV, Positive | splash10-0pxr-1219800000-b1461cae26afb8277a4b | 2017-09-01 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (2 TMS) - 70eV, Positive | splash10-03di-2011290000-9d901bc0dc4a11f91133 | 2017-10-06 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (TMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (TMS_1_3) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (TMS_2_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (TMS_2_3) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholic acid glycine conjugate GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Lithocholic acid glycine conjugate 10V, Positive-QTOF | splash10-00di-9003600000-1f7e0c91520f949fb8ec | 2016-08-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Lithocholic acid glycine conjugate 20V, Positive-QTOF | splash10-00di-9002000000-539de7f82b85a471b338 | 2016-08-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Lithocholic acid glycine conjugate 40V, Positive-QTOF | splash10-00di-9001000000-e53d44b83740a1b3b620 | 2016-08-01 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Lithocholic acid glycine conjugate 10V, Negative-QTOF | splash10-001i-0002900000-60e5b38e213f73a86996 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Lithocholic acid glycine conjugate 20V, Negative-QTOF | splash10-06z9-5107900000-ba230e9f34047158250a | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Lithocholic acid glycine conjugate 40V, Negative-QTOF | splash10-00dl-9102000000-859c6b8ef80fb6861c55 | 2016-08-03 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Lithocholic acid glycine conjugate 10V, Positive-QTOF | splash10-00lr-0002900000-29a47c1e04617905309c | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Lithocholic acid glycine conjugate 20V, Positive-QTOF | splash10-001m-4159200000-abff50ceed535345c378 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Lithocholic acid glycine conjugate 40V, Positive-QTOF | splash10-052g-9321000000-54d1ab9b1acc01fa59f1 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Lithocholic acid glycine conjugate 10V, Negative-QTOF | splash10-001i-0000900000-5cc587be0f34335a1e1c | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Lithocholic acid glycine conjugate 20V, Negative-QTOF | splash10-001i-0004900000-c9cb6871ee09ad0572f8 | 2021-09-24 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Lithocholic acid glycine conjugate 40V, Negative-QTOF | splash10-00e9-9002400000-076670a1180a04e9a7cc | 2021-09-24 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum |
|
|---|
| General References | - Tadano T, Kanoh M, Matsumoto M, Sakamoto K, Kamano T: Studies of serum and feces bile acids determination by gas chromatography-mass spectrometry. Rinsho Byori. 2006 Feb;54(2):103-10. [PubMed:16548228 ]
- Cowen AE, Korman MG, Hofmann AF, Cass OW, Coffin SB: Metabolism of lithocholate in healthy man. II. Enterohepatic circulation. Gastroenterology. 1975 Jul;69(1):67-76. [PubMed:1097294 ]
- Cowen AE, Korman MG, Hofmann AF, Cass OW: Metabolism of lethocholate in healthy man. I. Biotransformation and biliary excretion of intravenously administered lithocholate, lithocholylglycine, and their sulfates. Gastroenterology. 1975 Jul;69(1):59-66. [PubMed:1150035 ]
- Lee BL, New AL, Ong CN: Comparative analysis of conjugated bile acids in human serum using high-performance liquid chromatography and capillary electrophoresis. J Chromatogr B Biomed Sci Appl. 1997 Dec 19;704(1-2):35-42. [PubMed:9518169 ]
- Cowen AE, Korman MG, Hofmann AF, Thomas PJ: Metabolism of lithocholate in healthy man. III. Plasma disappearance of radioactivity after intravenous injection of labeled lithocholate and its derivatives. Gastroenterology. 1975 Jul;69(1):77-82. [PubMed:1150037 ]
- Goto T, Myint KT, Sato K, Wada O, Kakiyama G, Iida T, Hishinuma T, Mano N, Goto J: LC/ESI-tandem mass spectrometric determination of bile acid 3-sulfates in human urine 3beta-Sulfooxy-12alpha-hydroxy-5beta-cholanoic acid is an abundant nonamidated sulfate. J Chromatogr B Analyt Technol Biomed Life Sci. 2007 Feb 1;846(1-2):69-77. Epub 2006 Sep 1. [PubMed:16949895 ]
- St-Pierre MV, Kullak-Ublick GA, Hagenbuch B, Meier PJ: Transport of bile acids in hepatic and non-hepatic tissues. J Exp Biol. 2001 May;204(Pt 10):1673-86. [PubMed:11316487 ]
- Claudel T, Staels B, Kuipers F: The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol. 2005 Oct;25(10):2020-30. Epub 2005 Jul 21. [PubMed:16037564 ]
- Chiang JY: Bile acid regulation of hepatic physiology: III. Bile acids and nuclear receptors. Am J Physiol Gastrointest Liver Physiol. 2003 Mar;284(3):G349-56. [PubMed:12576301 ]
- Davis RA, Miyake JH, Hui TY, Spann NJ: Regulation of cholesterol-7alpha-hydroxylase: BAREly missing a SHP. J Lipid Res. 2002 Apr;43(4):533-43. [PubMed:11907135 ]
|
|---|