| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2023-02-21 17:14:29 UTC |
|---|
| HMDB ID | HMDB0000060 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Acetoacetic acid |
|---|
| Description | Acetoacetic acid (AcAc) is a weak organic acid that can be produced in the human liver under certain conditions of poor metabolism leading to excessive fatty acid breakdown (diabetes mellitus leading to diabetic ketoacidosis). It is then partially converted into acetone by decarboxylation and excreted either in urine or through respiration. Persistent mild hyperketonemia is a common finding in newborns. Ketone bodies serve as an indispensable source of energy for extrahepatic tissues, especially the brain and lung of developing rats. Another important function of ketone bodies is to provide acetoacetyl-CoA and acetyl-CoA for synthesis of cholesterol, fatty acids, and complex lipids. During the early postnatal period, acetoacetate and beta-hydroxybutyrate are preferred over glucose as substrates for synthesis of phospholipids and sphingolipids in accord with requirements for brain growth and myelination. Thus, during the first two weeks of postnatal development, when the accumulation of cholesterol and phospholipids accelerates, the proportion of ketone bodies incorporated into these lipids increases. On the other hand, an increased proportion of ketone bodies are utilized for cerebroside synthesis during the period of active myelination. In the lung, AcAc serves better than glucose as a precursor for the synthesis of lung phospholipids. The synthesized lipids, particularly dipalmityl phosphatidylcholine, are incorporated into surfactant, and thus have a potential role in supplying adequate surfactant lipids to maintain lung function during the early days of life (PMID: 3884391 ). The acid is also present in the metabolism of those undergoing starvation or prolonged physical exertion as part of gluconeogenesis. When ketone bodies are measured by way of urine concentration, acetoacetic acid, along with beta-hydroxybutyric acid or acetone, is what is detected. |
|---|
| Structure | InChI=1S/C4H6O3/c1-3(5)2-4(6)7/h2H2,1H3,(H,6,7) |
|---|
| Synonyms | | Value | Source |
|---|
| 3-Ketobutanoic acid | ChEBI | | 3-Ketobutyric acid | ChEBI | | 3-Oxobutanoic acid | ChEBI | | 3-Oxobutyric acid | ChEBI | | beta-Ketobutyric acid | ChEBI | | 3-Ketobutanoate | Generator | | 3-Ketobutyrate | Generator | | 3-Oxobutanoate | Generator | | 3-Oxobutyrate | Generator | | b-Ketobutyrate | Generator | | b-Ketobutyric acid | Generator | | beta-Ketobutyrate | Generator | | Β-ketobutyrate | Generator | | Β-ketobutyric acid | Generator | | Acetoacetate | Generator | | Acetoacetic acid, calcium salt | MeSH | | Acetoacetic acid, lithium salt | MeSH | | Acetoacetic acid, sodium salt | MeSH | | Oxobutyrate | MeSH | | Sodium acetoacetate | MeSH | | 3-oxo-Butanoate | HMDB | | 3-oxo-Butanoic acid | HMDB | | Diacetate | HMDB | | Diacetic acid | HMDB |
|
|---|
| Chemical Formula | C4H6O3 |
|---|
| Average Molecular Weight | 102.0886 |
|---|
| Monoisotopic Molecular Weight | 102.031694058 |
|---|
| IUPAC Name | 3-oxobutanoic acid |
|---|
| Traditional Name | acetoacetic acid |
|---|
| CAS Registry Number | 541-50-4 |
|---|
| SMILES | CC(=O)CC(O)=O |
|---|
| InChI Identifier | InChI=1S/C4H6O3/c1-3(5)2-4(6)7/h2H2,1H3,(H,6,7) |
|---|
| InChI Key | WDJHALXBUFZDSR-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as short-chain keto acids and derivatives. These are keto acids with an alkyl chain the contains less than 6 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Keto acids and derivatives |
|---|
| Sub Class | Short-chain keto acids and derivatives |
|---|
| Direct Parent | Short-chain keto acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Short-chain keto acid
- Beta-keto acid
- 1,3-dicarbonyl compound
- Beta-hydroxy ketone
- Ketone
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | |
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Acetoacetic acid,1TMS,isomer #1 | CC(=O)CC(=O)O[Si](C)(C)C | 1039.7 | Semi standard non polar | 33892256 | | Acetoacetic acid,1TMS,isomer #2 | CC(=CC(=O)O)O[Si](C)(C)C | 1168.5 | Semi standard non polar | 33892256 | | Acetoacetic acid,1TMS,isomer #3 | C=C(CC(=O)O)O[Si](C)(C)C | 1143.1 | Semi standard non polar | 33892256 | | Acetoacetic acid,2TMS,isomer #1 | CC(=CC(=O)O[Si](C)(C)C)O[Si](C)(C)C | 1234.5 | Semi standard non polar | 33892256 | | Acetoacetic acid,2TMS,isomer #1 | CC(=CC(=O)O[Si](C)(C)C)O[Si](C)(C)C | 1195.5 | Standard non polar | 33892256 | | Acetoacetic acid,2TMS,isomer #1 | CC(=CC(=O)O[Si](C)(C)C)O[Si](C)(C)C | 1227.9 | Standard polar | 33892256 | | Acetoacetic acid,2TMS,isomer #2 | C=C(CC(=O)O[Si](C)(C)C)O[Si](C)(C)C | 1187.7 | Semi standard non polar | 33892256 | | Acetoacetic acid,2TMS,isomer #2 | C=C(CC(=O)O[Si](C)(C)C)O[Si](C)(C)C | 1207.9 | Standard non polar | 33892256 | | Acetoacetic acid,2TMS,isomer #2 | C=C(CC(=O)O[Si](C)(C)C)O[Si](C)(C)C | 1275.0 | Standard polar | 33892256 | | Acetoacetic acid,1TBDMS,isomer #1 | CC(=O)CC(=O)O[Si](C)(C)C(C)(C)C | 1256.0 | Semi standard non polar | 33892256 | | Acetoacetic acid,1TBDMS,isomer #2 | CC(=CC(=O)O)O[Si](C)(C)C(C)(C)C | 1396.0 | Semi standard non polar | 33892256 | | Acetoacetic acid,1TBDMS,isomer #3 | C=C(CC(=O)O)O[Si](C)(C)C(C)(C)C | 1350.4 | Semi standard non polar | 33892256 | | Acetoacetic acid,2TBDMS,isomer #1 | CC(=CC(=O)O[Si](C)(C)C(C)(C)C)O[Si](C)(C)C(C)(C)C | 1638.4 | Semi standard non polar | 33892256 | | Acetoacetic acid,2TBDMS,isomer #1 | CC(=CC(=O)O[Si](C)(C)C(C)(C)C)O[Si](C)(C)C(C)(C)C | 1637.3 | Standard non polar | 33892256 | | Acetoacetic acid,2TBDMS,isomer #1 | CC(=CC(=O)O[Si](C)(C)C(C)(C)C)O[Si](C)(C)C(C)(C)C | 1558.1 | Standard polar | 33892256 | | Acetoacetic acid,2TBDMS,isomer #2 | C=C(CC(=O)O[Si](C)(C)C(C)(C)C)O[Si](C)(C)C(C)(C)C | 1617.7 | Semi standard non polar | 33892256 | | Acetoacetic acid,2TBDMS,isomer #2 | C=C(CC(=O)O[Si](C)(C)C(C)(C)C)O[Si](C)(C)C(C)(C)C | 1620.4 | Standard non polar | 33892256 | | Acetoacetic acid,2TBDMS,isomer #2 | C=C(CC(=O)O[Si](C)(C)C(C)(C)C)O[Si](C)(C)C(C)(C)C | 1584.7 | Standard polar | 33892256 |
|
|---|
| Spectra |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - Acetoacetic acid GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (Non-derivatized) | splash10-00kk-3940000000-008eb78d2ba3cfa053b4 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Acetoacetic acid GC-MS (1 MEOX; 1 TMS) | splash10-000i-9500000000-c3940e591f8aef9d6aac | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Acetoacetic acid GC-MS (1 MEOX; 1 TMS) | splash10-000i-9800000000-67a95675672c3f5447a7 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Acetoacetic acid GC-MS (2 TMS) | splash10-001i-4950000000-1778836b3908a79b814f | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Acetoacetic acid GC-MS (2 TMS) | splash10-001i-4950000000-4039bb48c98ec17b5c0c | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Acetoacetic acid GC-EI-TOF (Non-derivatized) | splash10-00kk-3940000000-008eb78d2ba3cfa053b4 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Acetoacetic acid GC-MS (Non-derivatized) | splash10-000i-9500000000-c3940e591f8aef9d6aac | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Acetoacetic acid GC-MS (Non-derivatized) | splash10-000i-9800000000-67a95675672c3f5447a7 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Acetoacetic acid GC-MS (Non-derivatized) | splash10-001i-4950000000-1778836b3908a79b814f | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - Acetoacetic acid GC-MS (Non-derivatized) | splash10-001i-4950000000-4039bb48c98ec17b5c0c | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Acetoacetic acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-0006-9000000000-9f6c0d100736f4d183c9 | 2016-09-22 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Acetoacetic acid GC-MS (1 TMS) - 70eV, Positive | splash10-0100-9600000000-470bf0676151478bca3e | 2017-10-06 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Acetoacetic acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Acetoacetic acid GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Acetoacetic acid GC-MS (TMS_1_3) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Acetoacetic acid GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Acetoacetic acid GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Acetoacetic acid GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Acetoacetic acid Quattro_QQQ 10V, N/A-QTOF (Annotated) | splash10-000i-9200000000-9b4ab7b6a41632cb125b | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Acetoacetic acid Quattro_QQQ 25V, N/A-QTOF (Annotated) | splash10-0udi-6900000000-cfab9d4cdb56acafe8cc | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Acetoacetic acid Quattro_QQQ 40V, N/A-QTOF (Annotated) | splash10-0pb9-9800000000-108aa5ef93455fb1ba28 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Acetoacetic acid , positive-QTOF | splash10-000i-9100000000-869e2338bbadb32272d4 | 2017-09-14 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Acetoacetic acid n/a 7V, negative-QTOF | splash10-0a4i-9000000000-3f979a5a82ec557199d2 | 2020-07-22 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Acetoacetic acid 20V, Negative-QTOF | splash10-000f-9000000000-2b35e5493fb76f714c22 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Acetoacetic acid 10V, Negative-QTOF | splash10-05fr-9000000000-d2734268ae2c1230e50f | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Acetoacetic acid 40V, Negative-QTOF | splash10-00di-9000000000-0d73b25163774c0d1eb1 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Acetoacetic acid 10V, Positive-QTOF | splash10-0f79-9400000000-081c4a1a05f5581a5ffb | 2016-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Acetoacetic acid 20V, Positive-QTOF | splash10-000l-9100000000-960a06fd90db43d3e0f6 | 2016-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Acetoacetic acid 40V, Positive-QTOF | splash10-0006-9000000000-bd091454ccc8f532b42e | 2016-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Acetoacetic acid 10V, Negative-QTOF | splash10-0zfr-9700000000-b01f47bb6cd4f1043983 | 2016-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Acetoacetic acid 20V, Negative-QTOF | splash10-0a4i-9100000000-d2649829e8a79045e73b | 2016-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Acetoacetic acid 40V, Negative-QTOF | splash10-0a4i-9000000000-1486ce68d51ba83ed499 | 2016-09-14 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Acetoacetic acid 10V, Negative-QTOF | splash10-0pb9-9400000000-2f187d15e3b22b7629d9 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Acetoacetic acid 20V, Negative-QTOF | splash10-0a4i-9100000000-1e5e193606848f9d64b4 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Acetoacetic acid 40V, Negative-QTOF | splash10-0a4l-9000000000-73a6b61cfeb4b8ae9612 | 2021-09-23 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Acetoacetic acid 10V, Positive-QTOF | splash10-0006-9000000000-de86214ccec455e1860b | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Acetoacetic acid 20V, Positive-QTOF | splash10-0006-9000000000-42af30a03d7e2d4c22fc | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Acetoacetic acid 40V, Positive-QTOF | splash10-0006-9000000000-6aa1f4078505fa3b0971 | 2021-09-25 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Experimental 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | 2021-10-10 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
|
|---|
| Biological Properties |
|---|
| Cellular Locations | - Cytoplasm
- Extracellular
- Mitochondria
- Peroxisome
|
|---|
| Biospecimen Locations | - Blood
- Cellular Cytoplasm
- Cerebrospinal Fluid (CSF)
- Feces
- Saliva
- Urine
|
|---|
| Tissue Locations | - Fibroblasts
- Leukocyte
- Liver
- Neuron
- Spleen
|
|---|
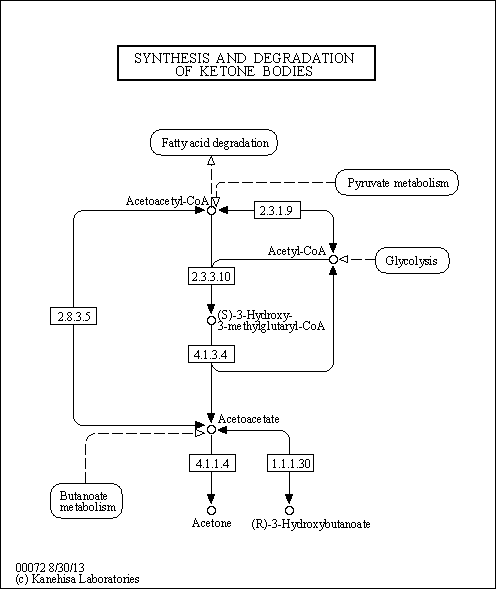
| Pathways | |
|---|
| Normal Concentrations |
|---|
| |
| Blood | Detected and Quantified | 21.0 (0.0-86.0) uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 37.75(25.96) uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | 30-1400 uM | Not Specified | Not Specified | Normal | | details | | Blood | Detected and Quantified | 0.0-86.0 uM | Adult (>18 years old) | Both | Normal | | details | | Blood | Detected and Quantified | <150 uM | Infant (0-1 year old) | Not Specified | Normal | | details | | Blood | Detected and Quantified | 20-90 uM | Infant (0-1 year old) | Not Specified | Normal | | details | | Blood | Detected and Quantified | 40.6 +/- 36.5 uM | Adult (>18 years old) | Both | Normal | | details | | Cellular Cytoplasm | Detected and Quantified | 1700 (1500-1900) uM | Adult (>18 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 26.2 +/- 12.3 uM | Adult (>18 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 12.0 +/- 14.0 uM | Adult (>18 years old) | Both | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 6.0 +/- 6.0 uM | Adult (>18 years old) | Not Specified | Normal | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 26.2 +/- 12.3 uM | Not Specified | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Not Specified | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Saliva | Detected and Quantified | 10.70 +/- 4.33 uM | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 0.15 (0.01-0.58) umol/mmol creatinine | Adult (>18 years old) | Male | Normal | | details | | Urine | Detected and Quantified | 0.20 (0.020-0.82) umol/mmol creatinine | Adult (>18 years old) | Female | Normal | | details | | Urine | Detected and Quantified | 11.1 (2.2-24.9) umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 18.64 +/- 20.321 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Normal | | details | | Urine | Detected and Quantified | 33.0(0.00-67.0) umol/mmol creatinine | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Normal | | details | | Urine | Detected and Quantified | 5-28 umol/mmol creatinine | Newborn (0-30 days old) | Both | Normal | | details |
|
|---|
| Abnormal Concentrations |
|---|
| |
| Blood | Detected and Quantified | 27.3 +/- 14.4 uM | Adult (>18 years old) | Both | Heart Transplant | | details | | Blood | Detected and Quantified | 19.56 +/- 10.63 uM | Adult (>18 years old) | Female | Pregnancy with fetuses with trisomy 18 | | details | | Blood | Detected and Quantified | 16.15 +/- 9.18 uM | Adult (>18 years old) | Female | Pregnancy | | details | | Blood | Detected and Quantified | 14.5 (5.1) uM | Adult (>18 years old) | Female | Early preeclampsia | | details | | Blood | Detected and Quantified | 18.8 (9.7) uM | Adult (>18 years old) | Female | Pregnancy | | details | | Blood | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Schizophrenia | | details | | Blood | Detected and Quantified | 3030.0 +/- 20.0 uM | Adult (>18 years old) | Both | Diabetes | | details | | Blood | Detected and Quantified | 14.33 (10.92) uM | Adult (>18 years old) | Female | Pregnancy with fetus having congenital heart defect | | details | | Blood | Detected and Quantified | 12.64 (8.21) uM | Adult (>18 years old) | Female | Pregnancy | | details | | Blood | Detected and Quantified | 18.9 (9.8) uM | Adult (>18 years old) | Female | Late-onset preeclampsia | | details | | Blood | Detected and Quantified | 16.5 (9.6) uM | Adult (>18 years old) | Female | Pregnancy | | details | | Blood | Detected and Quantified | 26.27(34.57) uM | Adult (>18 years old) | Both | Heart failure with reduced ejection fraction | | details | | Blood | Detected and Quantified | 100 uM | Newborn (0-30 days old) | Male | 2-Ketoglutarate dehydrogenase complex deficiency | | details | | Blood | Detected and Quantified | 50-1280 uM | Children (1-13 years old) | Male | 2-Ketoglutarate dehydrogenase complex deficiency | | details | | Blood | Detected and Quantified | 910 uM | Children (1-13 years old) | Male | Succinyl CoA: 3-ketoacid CoA transferase deficiency | | details | | Blood | Detected and Quantified | 1540 uM | Children (1-13 years old) | Male | Succinyl CoA: 3-ketoacid CoA transferase deficiency | | details | | Blood | Detected and Quantified | 360 uM | Infant (0-1 year old) | Male | Pyruvate dehydrogenase phosphatase deficiency | | details | | Blood | Detected and Quantified | 23.5 (24.3) uM | Adult (>18 years old) | Female | Down syndrome pregnancy | | details | | Blood | Detected and Quantified | 13.9 (7.3) uM | Adult (>18 years old) | Female | Pregnancy | | details | | Blood | Detected and Quantified | <30 uM | Children (1-13 years old) | Male | 3-Hydroxy-3-Methylglutaryl-CoA Synthase Deficiency | | details | | Cellular Cytoplasm | Detected and Quantified | 1000 (800-1200) uM | Adult (>18 years old) | Both | Anoxia | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 248 +/- 179 uM | Children (1-13 years old) | Not Specified | Glucose transporter type 1 deficiency syndrome | | details | | Cerebrospinal Fluid (CSF) | Detected and Quantified | 322.0 (240.0-404.0) uM | Adult (>18 years old) | Both | Meningitis | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Not Specified | Crohns disease | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Not Specified | Ulcerative colitis | | details | | Feces | Detected but not Quantified | Not Quantified | Children (6 - 18 years old) | Not Specified | Unclassified IBD | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Irritable Bowel Syndrome | | details | | Feces | Detected but not Quantified | Not Quantified | Adult (>18 years old) | Both | Colorectal cancer | | details | | Urine | Detected and Quantified | 1.6 (0.2-5.8) umol/mmol creatinine | Children (1-13 years old) | Both | Ketosis | | details | | Urine | Detected and Quantified | 0.1 (0-1.5) umol/mmol creatinine | Infant (0-1 year old) | Both | Ketosis | | details | | Urine | Detected and Quantified | 0.08 (0-1.5) umol/mmol creatinine | Newborn (0-30 days old) | Both | Ketosis | | details | | Urine | Detected and Quantified | 22.37 +/- 14.614 umol/mmol creatinine | Children (1 - 13 years old) | Not Specified | Eosinophilic esophagitis | | details | | Urine | Detected and Quantified | 237.0 umol/mmol creatinine | Adult (>18 years old) | Both | Diabetes | | details | | Urine | Detected and Quantified | 1.00 (0.00-2.00) umol/mmol creatinine | Adult (>18 years old) | Both | Ketosis | | details | | Urine | Detected and Quantified | 10025.00 (50.00-20000.00) umol/mmol creatinine | Children (1-13 years old) | Both | Ketosis | | details |
|
|---|
| Associated Disorders and Diseases |
|---|
| Disease References | | Diabetes mellitus type 2 |
|---|
- Tasker RC, Lutman D, Peters MJ: Hyperventilation in severe diabetic ketoacidosis. Pediatr Crit Care Med. 2005 Jul;6(4):405-11. [PubMed:15982426 ]
- Bales JR, Higham DP, Howe I, Nicholson JK, Sadler PJ: Use of high-resolution proton nuclear magnetic resonance spectroscopy for rapid multi-component analysis of urine. Clin Chem. 1984 Mar;30(3):426-32. [PubMed:6321058 ]
| | Schizophrenia |
|---|
- Cai HL, Li HD, Yan XZ, Sun B, Zhang Q, Yan M, Zhang WY, Jiang P, Zhu RH, Liu YP, Fang PF, Xu P, Yuan HY, Zhang XH, Hu L, Yang W, Ye HS: Metabolomic analysis of biochemical changes in the plasma and urine of first-episode neuroleptic-naive schizophrenia patients after treatment with risperidone. J Proteome Res. 2012 Aug 3;11(8):4338-50. doi: 10.1021/pr300459d. Epub 2012 Jul 26. [PubMed:22800120 ]
| | Early preeclampsia |
|---|
- Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K: Metabolomics and first-trimester prediction of early-onset preeclampsia. J Matern Fetal Neonatal Med. 2012 Oct;25(10):1840-7. doi: 10.3109/14767058.2012.680254. Epub 2012 Apr 28. [PubMed:22494326 ]
| | Pregnancy |
|---|
- Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K: Metabolomics and first-trimester prediction of early-onset preeclampsia. J Matern Fetal Neonatal Med. 2012 Oct;25(10):1840-7. doi: 10.3109/14767058.2012.680254. Epub 2012 Apr 28. [PubMed:22494326 ]
- Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K: First-trimester metabolomic detection of late-onset preeclampsia. Am J Obstet Gynecol. 2013 Jan;208(1):58.e1-7. doi: 10.1016/j.ajog.2012.11.003. Epub 2012 Nov 13. [PubMed:23159745 ]
- Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K: Metabolomic analysis for first-trimester Down syndrome prediction. Am J Obstet Gynecol. 2013 May;208(5):371.e1-8. doi: 10.1016/j.ajog.2012.12.035. Epub 2013 Jan 8. [PubMed:23313728 ]
- Bahado-Singh RO, Akolekar R, Chelliah A, Mandal R, Dong E, Kruger M, Wishart DS, Nicolaides K: Metabolomic analysis for first-trimester trisomy 18 detection. Am J Obstet Gynecol. 2013 Jul;209(1):65.e1-9. doi: 10.1016/j.ajog.2013.03.028. Epub 2013 Mar 25. [PubMed:23535240 ]
- Bahado-Singh RO, Ertl R, Mandal R, Bjorndahl TC, Syngelaki A, Han B, Dong E, Liu PB, Alpay-Savasan Z, Wishart DS, Nicolaides KH: Metabolomic prediction of fetal congenital heart defect in the first trimester. Am J Obstet Gynecol. 2014 Sep;211(3):240.e1-240.e14. doi: 10.1016/j.ajog.2014.03.056. Epub 2014 Apr 1. [PubMed:24704061 ]
| | Late-onset preeclampsia |
|---|
- Bahado-Singh RO, Akolekar R, Mandal R, Dong E, Xia J, Kruger M, Wishart DS, Nicolaides K: First-trimester metabolomic detection of late-onset preeclampsia. Am J Obstet Gynecol. 2013 Jan;208(1):58.e1-7. doi: 10.1016/j.ajog.2012.11.003. Epub 2012 Nov 13. [PubMed:23159745 ]
| | 2-Ketoglutarate dehydrogenase complex deficiency |
|---|
- Guffon N, Lopez-Mediavilla C, Dumoulin R, Mousson B, Godinot C, Carrier H, Collombet JM, Divry P, Mathieu M, Guibaud P: 2-Ketoglutarate dehydrogenase deficiency, a rare cause of primary hyperlactataemia: report of a new case. J Inherit Metab Dis. 1993;16(5):821-30. [PubMed:8295396 ]
| | 3-Hydroxy-3-Methylglutaryl-CoA Synthase Deficiency |
|---|
- Morris AA, Lascelles CV, Olpin SE, Lake BD, Leonard JV, Quant PA: Hepatic mitochondrial 3-hydroxy-3-methylglutaryl-coenzyme a synthase deficiency. Pediatr Res. 1998 Sep;44(3):392-6. doi: 10.1203/00006450-199809000-00021. [PubMed:9727719 ]
| | Ketoacidosis |
|---|
- Snyderman SE, Sansaricq C, Middleton B: Succinyl-CoA:3-ketoacid CoA-transferase deficiency. Pediatrics. 1998 Apr;101(4 Pt 1):709-11. [PubMed:9521962 ]
| | Pyruvate dehydrogenase phosphatase deficiency |
|---|
- Robinson BH, Sherwood WG: Pyruvate dehydrogenase phosphatase deficiency: a cause of congenital chronic lactic acidosis in infancy. Pediatr Res. 1975 Dec;9(12):935-9. doi: 10.1203/00006450-197512000-00015. [PubMed:172850 ]
| | Anoxia |
|---|
- Zupke C, Sinskey AJ, Stephanopoulos G: Intracellular flux analysis applied to the effect of dissolved oxygen on hybridomas. Appl Microbiol Biotechnol. 1995 Dec;44(1-2):27-36. [PubMed:8579834 ]
| | Glucose transporter type 1 deficiency syndrome |
|---|
- Klepper J, Diefenbach S, Kohlschutter A, Voit T: Effects of the ketogenic diet in the glucose transporter 1 deficiency syndrome. Prostaglandins Leukot Essent Fatty Acids. 2004 Mar;70(3):321-7. [PubMed:14769490 ]
| | Meningitis |
|---|
- Subramanian A, Gupta A, Saxena S, Gupta A, Kumar R, Nigam A, Kumar R, Mandal SK, Roy R: Proton MR CSF analysis and a new software as predictors for the differentiation of meningitis in children. NMR Biomed. 2005 Jun;18(4):213-25. [PubMed:15627241 ]
| | Irritable bowel syndrome |
|---|
- Hong YS, Hong KS, Park MH, Ahn YT, Lee JH, Huh CS, Lee J, Kim IK, Hwang GS, Kim JS: Metabonomic understanding of probiotic effects in humans with irritable bowel syndrome. J Clin Gastroenterol. 2011 May-Jun;45(5):415-25. doi: 10.1097/MCG.0b013e318207f76c. [PubMed:21494186 ]
| | Crohn's disease |
|---|
- Kolho KL, Pessia A, Jaakkola T, de Vos WM, Velagapudi V: Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J Crohns Colitis. 2017 Mar 1;11(3):321-334. doi: 10.1093/ecco-jcc/jjw158. [PubMed:27609529 ]
| | Ulcerative colitis |
|---|
- Kolho KL, Pessia A, Jaakkola T, de Vos WM, Velagapudi V: Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J Crohns Colitis. 2017 Mar 1;11(3):321-334. doi: 10.1093/ecco-jcc/jjw158. [PubMed:27609529 ]
| | Colorectal cancer |
|---|
- Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ: Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016 Mar 25;11(3):e0152126. doi: 10.1371/journal.pone.0152126. eCollection 2016. [PubMed:27015276 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
| | Ketosis |
|---|
- G.Frauendienst-Egger, Friedrich K. Trefz (2017). MetaGene: Metabolic & Genetic Information Center (MIC: http://www.metagene.de). METAGENE consortium.
- Blau N, Duran M, Blaskovics ME, Gibson KM (2003). Physician's Guide to the Laboratory Diagnosis of Metabolic Diseases. 2nd ed. Berlin, Germany, Springer, 2003. Springer.
| | Eosinophilic esophagitis |
|---|
- Slae, M., Huynh, H., Wishart, D.S. (2014). Analysis of 30 normal pediatric urine samples via NMR spectroscopy (unpublished work). NA.
|
|
|---|
| Associated OMIM IDs | - 125853 (Diabetes mellitus type 2)
- 181500 (Schizophrenia)
- 203740 (2-Ketoglutarate dehydrogenase complex deficiency)
- 605911 (3-Hydroxy-3-Methylglutaryl-CoA Synthase Deficiency)
- 245050 (Ketoacidosis)
- 608782 (Pyruvate dehydrogenase phosphatase deficiency)
- 606777 (Glucose transporter type 1 deficiency syndrome)
- 266600 (Crohn's disease)
- 114500 (Colorectal cancer)
- 610247 (Eosinophilic esophagitis)
|
|---|
| External Links |
|---|
| DrugBank ID | DB01762 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FooDB ID | FDB112157 |
|---|
| KNApSAcK ID | C00007458 |
|---|
| Chemspider ID | 94 |
|---|
| KEGG Compound ID | C00164 |
|---|
| BioCyc ID | 3-KETOBUTYRATE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Acetoacetic_acid |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 96 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15344 |
|---|
| Food Biomarker Ontology | Not Available |
|---|
| VMH ID | ACAC |
|---|
| MarkerDB ID | MDB00000030 |
|---|
| Good Scents ID | rw1468691 |
|---|
| References |
|---|
| Synthesis Reference | Lopez-Soriano, F. J.; Argiles, J. M. A simple method for the preparation of acetoacetate. Analytical Letters (1985), 18(B5), 589-92. |
|---|
| Material Safety Data Sheet (MSDS) | Download (PDF) |
|---|
| General References | - Nicholson JK, Foxall PJ, Spraul M, Farrant RD, Lindon JC: 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal Chem. 1995 Mar 1;67(5):793-811. [PubMed:7762816 ]
- Hoffmann GF, Meier-Augenstein W, Stockler S, Surtees R, Rating D, Nyhan WL: Physiology and pathophysiology of organic acids in cerebrospinal fluid. J Inherit Metab Dis. 1993;16(4):648-69. [PubMed:8412012 ]
- Subramanian A, Gupta A, Saxena S, Gupta A, Kumar R, Nigam A, Kumar R, Mandal SK, Roy R: Proton MR CSF analysis and a new software as predictors for the differentiation of meningitis in children. NMR Biomed. 2005 Jun;18(4):213-25. [PubMed:15627241 ]
- Zupke C, Sinskey AJ, Stephanopoulos G: Intracellular flux analysis applied to the effect of dissolved oxygen on hybridomas. Appl Microbiol Biotechnol. 1995 Dec;44(1-2):27-36. [PubMed:8579834 ]
- Heller MJ, Adams PW, Orosz CG: Evaluation of an automated method of percent reactive antibody determination. Hum Immunol. 1992 Nov;35(3):179-87. [PubMed:1293081 ]
- Tasker RC, Lutman D, Peters MJ: Hyperventilation in severe diabetic ketoacidosis. Pediatr Crit Care Med. 2005 Jul;6(4):405-11. [PubMed:15982426 ]
- Oligny LL, Lough J: Hepatic sinusoidal ectasia. Hum Pathol. 1992 Aug;23(8):953-6. [PubMed:1644440 ]
- Nicholson JK, O'Flynn MP, Sadler PJ, Macleod AF, Juul SM, Sonksen PH: Proton-nuclear-magnetic-resonance studies of serum, plasma and urine from fasting normal and diabetic subjects. Biochem J. 1984 Jan 15;217(2):365-75. [PubMed:6696735 ]
- Galey JB, Destree O, Dumats J, Genard S, Tachon P: Protection against oxidative damage by iron chelators: effect of lipophilic analogues and prodrugs of N,N'-bis(3,4,5-trimethoxybenzyl)ethylenediamine- N,N'-diacetic acid (OR10141). J Med Chem. 2000 Apr 6;43(7):1418-21. [PubMed:10753479 ]
- Saibara T, Onishi S, Sone J, Yamamoto N, Shimahara Y, Mori K, Ozawa K, Yamamoto Y: Arterial ketone body ratio as a possible indicator for liver transplantation in fulminant hepatic failure. Transplantation. 1991 Apr;51(4):782-6. [PubMed:2014530 ]
- Mahowald ML, Handwerger BS, Capertone EM Jr, Douglas SD: A comparative study of procedures for sheep erythrocyte-human-T-lymphocyte rosette formation. J Immunol Methods. 1977;15(3):239-45. [PubMed:404361 ]
- Sato T, Oouchi M, Nagakubo H, Chiba T, Ogawa S, Sato C, Sugimura K, Fukuda M: Effect of pravastatin on plasma ketone bodies in diabetics with hypercholesterolemia. Tohoku J Exp Med. 1998 May;185(1):25-9. [PubMed:9710942 ]
- Krejsa CM, Schieven GL: Detection of oxidative stress in lymphocytes using dichlorodihydrofluorescein diacetate. Methods Mol Biol. 2000;99:35-47. [PubMed:10909075 ]
- Fritzsche I, Buhrdel P, Melcher R, Bohme HJ: Stability of ketone bodies in serum in dependence on storage time and storage temperature. Clin Lab. 2001;47(7-8):399-403. [PubMed:11499803 ]
- Polsky-Fisher SL, Cao H, Lu P, Gibson CR: Effect of cytochromes P450 chemical inhibitors and monoclonal antibodies on human liver microsomal esterase activity. Drug Metab Dispos. 2006 Aug;34(8):1361-6. Epub 2006 May 23. [PubMed:16720683 ]
- Fulop M, Murthy V, Michilli A, Nalamati J, Qian Q, Saitowitz A: Serum beta-hydroxybutyrate measurement in patients with uncontrolled diabetes mellitus. Arch Intern Med. 1999 Feb 22;159(4):381-4. [PubMed:10030312 ]
- Tanaka Y, Ohdan H, Onoe T, Mitsuta H, Tashiro H, Itamoto T, Asahara T: Low incidence of acute rejection after living-donor liver transplantation: immunologic analyses by mixed lymphocyte reaction using a carboxyfluorescein diacetate succinimidyl ester labeling technique. Transplantation. 2005 May 15;79(9):1262-7. [PubMed:15880082 ]
- Bales JR, Higham DP, Howe I, Nicholson JK, Sadler PJ: Use of high-resolution proton nuclear magnetic resonance spectroscopy for rapid multi-component analysis of urine. Clin Chem. 1984 Mar;30(3):426-32. [PubMed:6321058 ]
- de Araujo Burgos MG, Bion FM, Campos F: [Lactation and alcohol: clinical and nutritional effects]. Arch Latinoam Nutr. 2004 Mar;54(1):25-35. [PubMed:15332353 ]
- Walker V, Mills GA, Mellor JM, Langley GJ, Farrant RD: A novel pyrroline-5-carboxylic acid and acetoacetic acid adduct in hyperprolinaemia type II. Clin Chim Acta. 2003 May;331(1-2):7-17. [PubMed:12691858 ]
- Yeh YY, Sheehan PM: Preferential utilization of ketone bodies in the brain and lung of newborn rats. Fed Proc. 1985 Apr;44(7):2352-8. [PubMed:3884391 ]
|
|---|