| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:49:00 UTC |
|---|
| HMDB ID | HMDB0000235 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Thiamine |
|---|
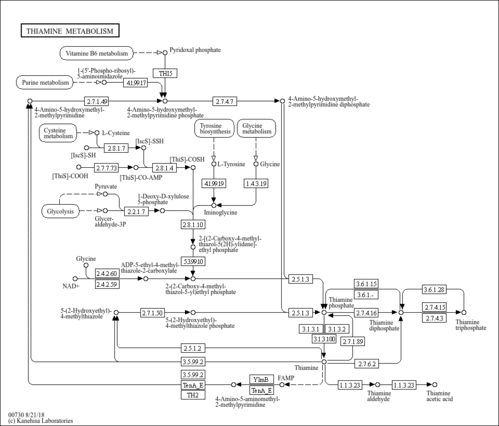
| Description | Thiamine, also known as aneurin or vitamin B1, belongs to the class of organic compounds known as thiamines. Thiamines are compounds containing a thiamine moiety, which is structurally characterized by a 3-[(4-Amino-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-thiazol-5-yl backbone. Thiamine exists in all living species, ranging from bacteria to plants to humans. Thiamine biosynthesis occurs in bacteria, some protozoans, plants, and fungi. Thiamine is a vitamin and an essential nutrient meaning the body cannot synthesize it, and it must be obtained from the diet. It is soluble in water and insoluble in alcohol. Thiamine decomposes if heated. Thiamine was first discovered in 1897 by Umetaro Suzuki in Japan when researching how rice bran cured patients of Beriberi. Thiamine was the first B vitamin to be isolated in 1926 and was first made in 1936. Thiamine plays a key role in intracellular glucose metabolism and it is thought that thiamine inhibits the effect of glucose and insulin on arterial smooth muscle cell proliferation. Thiamine plays an important role in helping the body convert carbohydrates and fat into energy. It is essential for normal growth and development and helps to maintain proper functioning of the heart and the nervous and digestive systems. Thiamine cannot be stored in the body; however, once absorbed, the vitamin is concentrated in muscle tissue. Thiamine has antioxidant, erythropoietic, cognition-and mood-modulatory, antiatherosclerotic, putative ergogenic, and detoxification activities. Natural derivatives of thiamine, such as thiamine monophosphate (ThMP), thiamine diphosphate (ThDP), also sometimes called thiamine pyrophosphate (TPP), thiamine triphosphate (ThTP), and adenosine thiamine triphosphate (AThTP), act as coenzymes in addition to performing unique biological functions. Thiamine deficiency can lead to beriberi, Wernicke-Korsakoff syndrome, optic neuropathy, Leigh's disease, African seasonal ataxia (or Nigerian seasonal ataxia), and central pontine myelinolysis. In Western countries, thiamine deficiency is seen mainly in chronic alcoholism. Thiamine supplements or thiamine therapy can be used for the treatment of a number of disorders including thiamine and niacin deficiency states, Korsakov's alcoholic psychosis, Wernicke-Korsakov syndrome, delirium, and peripheral neuritis. In humans, thiamine is involved in the metabolic disorder called 2-methyl-3-hydroxybutyryl-CoA dehydrogenase deficiency. Outside of the human body, Thiamine is found in high quantities in whole grains, legumes, pork, fruits, and yeast and fish. Grain processing removes much of the thiamine content in grains, so in many countries cereals and flours are enriched with thiamine. |
|---|
| Structure | CC1=C(CCO)SC=[N+]1CC1=CN=C(C)N=C1N InChI=1S/C12H17N4OS/c1-8-11(3-4-17)18-7-16(8)6-10-5-14-9(2)15-12(10)13/h5,7,17H,3-4,6H2,1-2H3,(H2,13,14,15)/q+1 |
|---|
| Synonyms | | Value | Source |
|---|
| 3-(4-AMINO-2-methyl-pyrimidin-5-ylmethyl)-5-(2-hydroxy-ethyl)-4-methyl-thiazol-3-ium | ChEBI | | Aneurin | ChEBI | | Antiberiberi factor | ChEBI | | Thiamin | ChEBI | | Thiamine(1+) ion | ChEBI | | Thiaminium | ChEBI | | Vitamin b1 | ChEBI | | Apate drops | HMDB | | Beatine | HMDB | | Bedome | HMDB | | Begiolan | HMDB | | Benerva | HMDB | | Bequin | HMDB | | Berin | HMDB | | Betalin S | HMDB | | Betaxin | HMDB | | Bethiazine | HMDB | | Beuion | HMDB | | Bevitex | HMDB | | Bevitine | HMDB | | Bewon | HMDB | | Biamine | HMDB | | Bithiamin | HMDB | | Biuno | HMDB | | Bivatin | HMDB | | Bivita | HMDB | | Cernevit-12 | HMDB | | Clotiamina | HMDB | | Eskapen | HMDB | | Eskaphen | HMDB | | Hybee | HMDB | | Lixa-beta | HMDB | | Metabolin | HMDB | | Slowten | HMDB | | THD | HMDB | | Thiadoxine | HMDB | | Thiaminal | HMDB | | Thiamol | HMDB | | Thiavit | HMDB | | Tiamidon | HMDB | | Tiaminal | HMDB | | Trophite | HMDB | | Vetalin S | HMDB | | VIB | HMDB | | Vinothiam | HMDB | | Vitaneuron | HMDB | | Mononitrate, thiamine | MeSH, HMDB | | Thiamine mononitrate | MeSH, HMDB | | Vitamin b 1 | MeSH, HMDB |
|
|---|
| Chemical Formula | C12H17N4OS |
|---|
| Average Molecular Weight | 265.355 |
|---|
| Monoisotopic Molecular Weight | 265.112306876 |
|---|
| IUPAC Name | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-hydroxyethyl)-4-methyl-1,3-thiazol-3-ium |
|---|
| Traditional Name | thiamine |
|---|
| CAS Registry Number | 70-16-6 |
|---|
| SMILES | CC1=C(CCO)SC=[N+]1CC1=CN=C(C)N=C1N |
|---|
| InChI Identifier | InChI=1S/C12H17N4OS/c1-8-11(3-4-17)18-7-16(8)6-10-5-14-9(2)15-12(10)13/h5,7,17H,3-4,6H2,1-2H3,(H2,13,14,15)/q+1 |
|---|
| InChI Key | JZRWCGZRTZMZEH-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as thiamines. Thiamines are compounds containing a thiamine moiety, which is structurally characterized by a 3-[(4-Amino-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-thiazol-5-yl backbone. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Diazines |
|---|
| Sub Class | Pyrimidines and pyrimidine derivatives |
|---|
| Direct Parent | Thiamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Thiamine
- 4,5-disubstituted 1,3-thiazole
- Aminopyrimidine
- Imidolactam
- Azole
- Thiazole
- Heteroaromatic compound
- Azacycle
- Alcohol
- Organopnictogen compound
- Primary amine
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Organic oxygen compound
- Hydrocarbon derivative
- Amine
- Organic cation
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | |
|---|
| Disposition | Biological locationRoute of exposureSourceExogenousFood- Food (HMDB: HMDB0000235)
Animal originVegetableFruitHerb and spiceNutCereal and cereal productPulseGourdSoyTeaBeverageAquatic originEggConfectioneryMilk and milk productOther milk productFermented milkFermented milk productUnfermented milk- Milk (Other mammals) (FooDB: FOOD00690)
- Milk (Human) (FooDB: FOOD00666)
- Milk (Cow) (FooDB: FOOD00618)
- Cow milk, pasteurized, vitamin A + D added, 0% fat (FooDB: FOOD00889)
- Cow milk, pasteurized, vitamin A + D added, 1% fat (FooDB: FOOD00890)
- Cow milk, pasteurized, vitamin A + D added, 2% fat (FooDB: FOOD00891)
- Cow milk, pasteurized, vitamin D added, 3.25% fat (FooDB: FOOD00892)
- Milk and milk products (FooDB: FOOD00863)
Fat and oilCocoa and cocoa productBaking goodBaby foodUnclassified food or beverageDishCoffee and coffee productSnack
Synthetic Endogenous |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 248 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 500 mg/mL | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections| Adduct Type | Data Source | CCS Value (Å2) | Reference |
|---|
| [M-H]- | MetCCS_train_neg | 160.3 | 30932474 | | [M+H]+ | MetCCS_test_pos | 162.373 | 30932474 |
|
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Thiamine,1TMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C)=C2C)C(N)=N1 | 2471.7 | Semi standard non polar | 33892256 | | Thiamine,1TMS,isomer #2 | CC1=NC=C(C[N+]2=CSC(CCO)=C2C)C(N[Si](C)(C)C)=N1 | 2549.4 | Semi standard non polar | 33892256 | | Thiamine,2TMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C)=C2C)C(N[Si](C)(C)C)=N1 | 2502.4 | Semi standard non polar | 33892256 | | Thiamine,2TMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C)=C2C)C(N[Si](C)(C)C)=N1 | 2467.2 | Standard non polar | 33892256 | | Thiamine,2TMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C)=C2C)C(N[Si](C)(C)C)=N1 | 3264.5 | Standard polar | 33892256 | | Thiamine,2TMS,isomer #2 | CC1=NC=C(C[N+]2=CSC(CCO)=C2C)C(N([Si](C)(C)C)[Si](C)(C)C)=N1 | 2540.1 | Semi standard non polar | 33892256 | | Thiamine,2TMS,isomer #2 | CC1=NC=C(C[N+]2=CSC(CCO)=C2C)C(N([Si](C)(C)C)[Si](C)(C)C)=N1 | 2549.2 | Standard non polar | 33892256 | | Thiamine,2TMS,isomer #2 | CC1=NC=C(C[N+]2=CSC(CCO)=C2C)C(N([Si](C)(C)C)[Si](C)(C)C)=N1 | 3162.5 | Standard polar | 33892256 | | Thiamine,3TMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C)=C2C)C(N([Si](C)(C)C)[Si](C)(C)C)=N1 | 2514.5 | Semi standard non polar | 33892256 | | Thiamine,3TMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C)=C2C)C(N([Si](C)(C)C)[Si](C)(C)C)=N1 | 2583.7 | Standard non polar | 33892256 | | Thiamine,3TMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C)=C2C)C(N([Si](C)(C)C)[Si](C)(C)C)=N1 | 2946.7 | Standard polar | 33892256 | | Thiamine,1TBDMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C(C)(C)C)=C2C)C(N)=N1 | 2740.7 | Semi standard non polar | 33892256 | | Thiamine,1TBDMS,isomer #2 | CC1=NC=C(C[N+]2=CSC(CCO)=C2C)C(N[Si](C)(C)C(C)(C)C)=N1 | 2747.4 | Semi standard non polar | 33892256 | | Thiamine,2TBDMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C(C)(C)C)=C2C)C(N[Si](C)(C)C(C)(C)C)=N1 | 2905.4 | Semi standard non polar | 33892256 | | Thiamine,2TBDMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C(C)(C)C)=C2C)C(N[Si](C)(C)C(C)(C)C)=N1 | 2897.6 | Standard non polar | 33892256 | | Thiamine,2TBDMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C(C)(C)C)=C2C)C(N[Si](C)(C)C(C)(C)C)=N1 | 3365.0 | Standard polar | 33892256 | | Thiamine,2TBDMS,isomer #2 | CC1=NC=C(C[N+]2=CSC(CCO)=C2C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=N1 | 2927.6 | Semi standard non polar | 33892256 | | Thiamine,2TBDMS,isomer #2 | CC1=NC=C(C[N+]2=CSC(CCO)=C2C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=N1 | 2959.6 | Standard non polar | 33892256 | | Thiamine,2TBDMS,isomer #2 | CC1=NC=C(C[N+]2=CSC(CCO)=C2C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=N1 | 3242.0 | Standard polar | 33892256 | | Thiamine,3TBDMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C(C)(C)C)=C2C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=N1 | 3094.1 | Semi standard non polar | 33892256 | | Thiamine,3TBDMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C(C)(C)C)=C2C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=N1 | 3195.6 | Standard non polar | 33892256 | | Thiamine,3TBDMS,isomer #1 | CC1=NC=C(C[N+]2=CSC(CCO[Si](C)(C)C(C)(C)C)=C2C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=N1 | 3187.4 | Standard polar | 33892256 |
|
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - Thiamine GC-MS (Non-derivatized) - 70eV, Positive | splash10-00e9-3890000000-9f8b525433ab279ad512 | 2016-09-22 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Thiamine GC-MS (1 TMS) - 70eV, Positive | splash10-00di-9442000000-05ab93d3afb7b6f538e9 | 2017-10-06 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Thiamine GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Thiamine GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Thiamine GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Thiamine GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine Quattro_QQQ 10V, N/A-QTOF (Annotated) | splash10-000i-0109000000-48864f31ba475645654b | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine Quattro_QQQ 25V, N/A-QTOF (Annotated) | splash10-05o1-9200000000-2ec0f3e9e364cffde16e | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine Quattro_QQQ 40V, N/A-QTOF (Annotated) | splash10-0aou-9000000000-0184506492d9f5b68b97 | 2012-07-24 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative-QTOF | splash10-03di-0090000000-872150be83cbd75d8ecf | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative-QTOF | splash10-0002-0930000000-ecf71c75bdea859139d5 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative-QTOF | splash10-0002-0900000000-4d9113fdc633fb1ccbb5 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative-QTOF | splash10-0002-0900000000-4a8016299623af0c3027 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative-QTOF | splash10-0002-1900000000-773912c9f325c419c77d | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive-QTOF | splash10-014i-0390000000-95f3ca57c95542bdc7e7 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive-QTOF | splash10-00di-0900000000-e079d1fc1e06396dc524 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive-QTOF | splash10-00di-0900000000-ebb16d4f204f291b9a3a | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive-QTOF | splash10-00e9-4900000000-ad4d4779d76b9edb7ec5 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive-QTOF | splash10-001i-9600000000-b631eed5be831527fc17 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive-QTOF | splash10-00di-0900000000-50f2f613f576814eb627 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive-QTOF | splash10-002b-0900000000-2f184bbfec6bc0ef00c6 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive-QTOF | splash10-001i-9000000000-35bc744495a2570a565f | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive-QTOF | splash10-00di-1900000000-d8a7ac327273c32b4524 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative-QTOF | splash10-0002-0920000000-ca80ef6513c61be5b5e9 | 2012-08-31 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Thiamine LC-ESI-QQ , negative-QTOF | splash10-03di-0090000000-872150be83cbd75d8ecf | 2017-09-14 | HMDB team, MONA | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Thiamine 10V, Positive-QTOF | splash10-014i-0190000000-183af2449d1439af9825 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Thiamine 20V, Positive-QTOF | splash10-0a4i-0090000000-2230ebc7c6b73192c26f | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Thiamine 40V, Positive-QTOF | splash10-0002-7920000000-75670df9647a475ca92c | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Thiamine 10V, Negative-QTOF | splash10-03di-3090000000-d0ff9a733629a86b75c7 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Thiamine 20V, Negative-QTOF | splash10-03dj-3690000000-db3f204af490d7e1722e | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - Thiamine 40V, Negative-QTOF | splash10-0005-9000000000-5196f2b4daab995967bf | 2016-09-12 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Experimental 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | 2012-12-04 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-29 | Wishart Lab | View Spectrum | | Experimental 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | 2021-10-10 | Wishart Lab | View Spectrum | | Experimental 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | 2012-12-05 | Wishart Lab | View Spectrum |
|
|---|
| Disease References | | Hemodialysis |
|---|
- Hung SC, Hung SH, Tarng DC, Yang WC, Chen TW, Huang TP: Thiamine deficiency and unexplained encephalopathy in hemodialysis and peritoneal dialysis patients. Am J Kidney Dis. 2001 Nov;38(5):941-7. [PubMed:11684545 ]
| | Friedreich's ataxia |
|---|
- Pedraza OL, Botez MI: Thiamine status in inherited degenerative ataxias. J Neurol Neurosurg Psychiatry. 1992 Feb;55(2):136-7. [PubMed:1538220 ]
- Botez MI, Young SN: Biogenic amine metabolites and thiamine in cerebrospinal fluid in heredo-degenerative ataxias. Can J Neurol Sci. 2001 May;28(2):134-40. [PubMed:11383938 ]
| | Olivopontocerebral atrophy |
|---|
- Pedraza OL, Botez MI: Thiamine status in inherited degenerative ataxias. J Neurol Neurosurg Psychiatry. 1992 Feb;55(2):136-7. [PubMed:1538220 ]
- Botez MI, Young SN: Biogenic amine metabolites and thiamine in cerebrospinal fluid in heredo-degenerative ataxias. Can J Neurol Sci. 2001 May;28(2):134-40. [PubMed:11383938 ]
| | Alcoholism |
|---|
- Dastur DK, Santhadevi N, Quadros EV, Avari FC, Wadia NH, Desai MN, Bharucha EP: The B-vitamins in malnutrition with alcoholism. A model of intervitamin relationships. Br J Nutr. 1976 Sep;36(2):143-59. [PubMed:182198 ]
| | Hereditary spastic paraplegia |
|---|
- Botez MI, Young SN: Biogenic amine metabolites and thiamine in cerebrospinal fluid in heredo-degenerative ataxias. Can J Neurol Sci. 2001 May;28(2):134-40. [PubMed:11383938 ]
| | Colorectal cancer |
|---|
- Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, Hayes RB, Goedert JJ: Fecal Microbiota, Fecal Metabolome, and Colorectal Cancer Interrelations. PLoS One. 2016 Mar 25;11(3):e0152126. doi: 10.1371/journal.pone.0152126. eCollection 2016. [PubMed:27015276 ]
- Goedert JJ, Sampson JN, Moore SC, Xiao Q, Xiong X, Hayes RB, Ahn J, Shi J, Sinha R: Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis. 2014 Sep;35(9):2089-96. doi: 10.1093/carcin/bgu131. Epub 2014 Jul 18. [PubMed:25037050 ]
|
|
|---|
| General References | - Dutta B, Huang W, Molero M, Kekuda R, Leibach FH, Devoe LD, Ganapathy V, Prasad PD: Cloning of the human thiamine transporter, a member of the folate transporter family. J Biol Chem. 1999 Nov 5;274(45):31925-9. [PubMed:10542220 ]
- Bellazzi R, Guglielmann R, Ironi L, Patrini C: A hybrid input-output approach to model metabolic systems: an application to intracellular thiamine kinetics. J Biomed Inform. 2001 Aug;34(4):221-48. [PubMed:11977806 ]
- Pietrzak I, Baczyk K: Comparison of the thiamine level in blood and erythrocyte transketolase activity in hemodialyzed and nondialyzed patients during recombinant human erythropoietin therapy. Miner Electrolyte Metab. 1997;23(3-6):277-82. [PubMed:9387133 ]
- Singleton CK, Martin PR: Molecular mechanisms of thiamine utilization. Curr Mol Med. 2001 May;1(2):197-207. [PubMed:11899071 ]
- Sato Y, Nakagawa M, Higuchi I, Osame M, Naito E, Oizumi K: Mitochondrial myopathy and familial thiamine deficiency. Muscle Nerve. 2000 Jul;23(7):1069-75. [PubMed:10883001 ]
- Mastrogiacoma F, Bettendorff L, Grisar T, Kish SJ: Brain thiamine, its phosphate esters, and its metabolizing enzymes in Alzheimer's disease. Ann Neurol. 1996 May;39(5):585-91. [PubMed:8619543 ]
- Molina JA, Jimenez-Jimenez FJ, Hernanz A, Fernandez-Vivancos E, Medina S, de Bustos F, Gomez-Escalonilla C, Sayed Y: Cerebrospinal fluid levels of thiamine in patients with Alzheimer's disease. J Neural Transm (Vienna). 2002 Jul;109(7-8):1035-44. [PubMed:12111441 ]
- Pietrzak I, Baczyk K, Kubiak W: Recombinant human erythropoietin administration improves thiamine content in blood and erythrocytes transketolase activity in pre-dialyzed patients. Ann Univ Mariae Curie Sklodowska Med. 1994;48 Suppl 3:29-37. [PubMed:8192530 ]
- Valerio G, Franzese A, Poggi V, Patrini C, Laforenza U, Tenore A: Lipophilic thiamine treatment in long-standing insulin-dependent diabetes mellitus. Acta Diabetol. 1999 Jun;36(1-2):73-6. [PubMed:10436256 ]
- Herve C, Beyne P, Delacoux E: Determination of thiamine and its phosphate esters in human erythrocytes by high-performance liquid chromatography with isocratic elution. J Chromatogr B Biomed Appl. 1994 Mar 4;653(2):217-20. [PubMed:8205249 ]
- Losa R, Sierra MI, Fernandez A, Blanco D, Buesa JM: Determination of thiamine and its phosphorylated forms in human plasma, erythrocytes and urine by HPLC and fluorescence detection: a preliminary study on cancer patients. J Pharm Biomed Anal. 2005 Apr 29;37(5):1025-9. [PubMed:15862682 ]
- Pietrzak I, Baczyk K, Mlynarczyk M: [The influence of intermittent peritoneal dialysis on free and total thiamine concentration in plasma and erythrocytes of patients with end stage renal disease]. Pol Arch Med Wewn. 1994;92 Spec No:31-6. [PubMed:7731897 ]
- Pedraza OL, Botez MI: Thiamine status in inherited degenerative ataxias. J Neurol Neurosurg Psychiatry. 1992 Feb;55(2):136-7. [PubMed:1538220 ]
- Vinogradov VV, Tarasov IuA, Tishin VS, Bogdanovich VI, Spas VV: [Thiamine prevention of the corticosteroid reaction afer surgery]. Probl Endokrinol (Mosk). 1981 May-Jun;27(3):11-6. [PubMed:7291135 ]
- Tanaka T, Sohmiya K, Kono T, Terasaki F, Horie R, Ohkaru Y, Muramatsu M, Takai S, Miyazaki M, Kitaura Y: Thiamine attenuates the hypertension and metabolic abnormalities in CD36-defective SHR: uncoupling of glucose oxidation from cellular entry accompanied with enhanced protein O-GlcNAcylation in CD36 deficiency. Mol Cell Biochem. 2007 May;299(1-2):23-35. [PubMed:16645728 ]
- Bettendorff L, Mastrogiacomo F, Kish SJ, Grisar T: Thiamine, thiamine phosphates, and their metabolizing enzymes in human brain. J Neurochem. 1996 Jan;66(1):250-8. [PubMed:8522961 ]
- Lee DC, Chu J, Satz W, Silbergleit R: Low plasma thiamine levels in elder patients admitted through the emergency department. Acad Emerg Med. 2000 Oct;7(10):1156-9. [PubMed:11015250 ]
- Shimon I, Almog S, Vered Z, Seligmann H, Shefi M, Peleg E, Rosenthal T, Motro M, Halkin H, Ezra D: Improved left ventricular function after thiamine supplementation in patients with congestive heart failure receiving long-term furosemide therapy. Am J Med. 1995 May;98(5):485-90. [PubMed:7733128 ]
- Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. [PubMed:32033212 ]
|
|---|