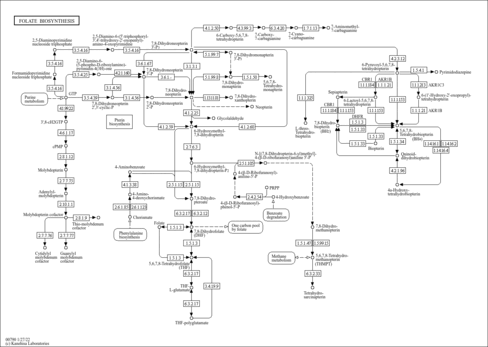
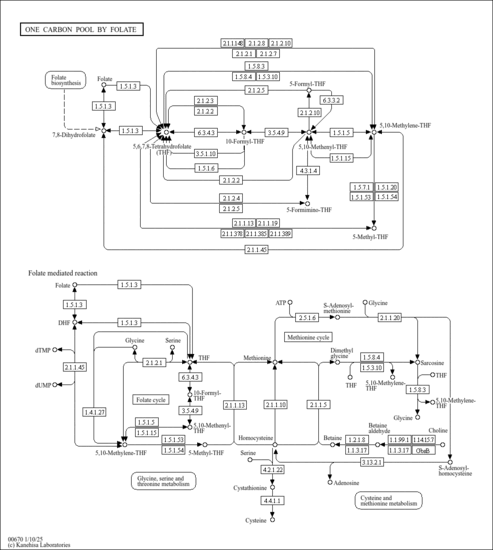
| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Dihydrofolic acid,1TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)C(=O)O | 4626.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,1TMS,isomer #2 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1 | 4621.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,1TMS,isomer #3 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)[NH]1 | 4796.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,1TMS,isomer #4 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1[NH]C(N)=NC2=O | 4528.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,1TMS,isomer #5 | C[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4664.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,1TMS,isomer #6 | C[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 4616.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,1TMS,isomer #7 | C[Si](C)(C)N1C(N)=NC(=O)C2=C1NCC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1)=N2 | 4639.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 4450.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #10 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1 | 4454.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #11 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1 | 4306.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #12 | C[Si](C)(C)N(C1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)[NH]1)[Si](C)(C)C | 4613.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #13 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C | 4622.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #14 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1)=N2 | 4506.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #15 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4630.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #16 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)=N2)[NH]1 | 4581.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #17 | C[Si](C)(C)N(CC1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4342.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #18 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C2)=NC2=C1[NH]C(N)=NC2=O | 4274.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #19 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1N([Si](C)(C)C)C(N)=NC2=O | 4381.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #2 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)=N2)[NH]1 | 4564.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #20 | C[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4469.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #21 | C[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4394.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #22 | C[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 4425.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)[Si](C)(C)C | 4407.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 4444.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #5 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)C(=O)O | 4455.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)C(=O)O | 4305.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)=N2)[NH]1 | 4578.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #8 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)[Si](C)(C)C | 4435.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TMS,isomer #9 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1 | 4434.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #1 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)=N2)[NH]1 | 4435.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #1 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)=N2)[NH]1 | 4082.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #1 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)=N2)[NH]1 | 7544.3 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #10 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)=N2)[NH]1 | 4386.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #10 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)=N2)[NH]1 | 4058.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #10 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)=N2)[NH]1 | 7521.2 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #11 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4211.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #11 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3949.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #11 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 6702.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #12 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 4280.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #12 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 3960.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #12 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 7024.2 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #13 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 4113.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #13 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 4016.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #13 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 6704.3 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #14 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 4305.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #14 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 4032.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #14 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 6886.3 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #15 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)C(=O)O | 4145.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #15 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)C(=O)O | 4074.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #15 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)C(=O)O | 6610.1 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #16 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)C(=O)O | 4255.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #16 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)C(=O)O | 4105.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #16 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)C(=O)O | 6834.0 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #17 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1 | 4434.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #17 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1 | 4107.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #17 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1 | 7190.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #18 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4473.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #18 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4088.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #18 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 7350.4 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #19 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C1)=N2 | 4345.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #19 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C1)=N2 | 4105.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #19 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C1)=N2 | 7052.0 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)[Si](C)(C)C | 4309.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)[Si](C)(C)C | 3903.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)[Si](C)(C)C | 6873.4 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #20 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4414.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #20 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4069.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #20 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 7328.7 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #21 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)[NH]1 | 4401.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #21 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)[NH]1 | 4007.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #21 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)[NH]1 | 7474.0 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #22 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4226.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #22 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3908.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #22 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 6651.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #23 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 4284.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #23 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 3914.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #23 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 6973.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #24 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 4133.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #24 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 3970.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #24 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 6655.5 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #25 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1 | 4278.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #25 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1 | 3977.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #25 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1 | 6803.6 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #26 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1 | 4136.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #26 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1 | 4029.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #26 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1 | 6531.5 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #27 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1 | 4244.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #27 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1 | 4057.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #27 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1 | 6754.3 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #28 | C[Si](C)(C)N(C1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C)[Si](C)(C)C | 4522.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #28 | C[Si](C)(C)N(C1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C)[Si](C)(C)C | 4166.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #28 | C[Si](C)(C)N(C1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C)[Si](C)(C)C | 7271.2 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #29 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4364.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #29 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4210.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #29 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 6954.1 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 4294.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 3953.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 6729.2 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #30 | C[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4478.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #30 | C[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4169.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #30 | C[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 7066.4 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #31 | C[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 4397.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #31 | C[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 4092.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #31 | C[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 7195.9 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #32 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C | 4435.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #32 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C | 4233.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #32 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C | 6931.6 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #33 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4487.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #33 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4145.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #33 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 7199.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #34 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4435.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #34 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4078.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #34 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 7338.5 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #35 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1)[Si](C)(C)C)=N2 | 4370.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #35 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1)[Si](C)(C)C)=N2 | 4179.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #35 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1)[Si](C)(C)C)=N2 | 6943.9 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #36 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1)=N2 | 4321.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #36 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1)=N2 | 4111.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #36 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1)=N2 | 7044.7 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #37 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4376.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #37 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4048.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #37 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 7300.5 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #38 | C[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4259.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #38 | C[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4130.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #38 | C[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 6626.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #39 | C[Si](C)(C)N(CC1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4103.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #39 | C[Si](C)(C)N(CC1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4037.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #39 | C[Si](C)(C)N(CC1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C)C1)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 6511.2 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 4343.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 3967.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 7034.0 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #40 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C2)=NC2=C1N([Si](C)(C)C)C(N)=NC2=O | 4198.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #40 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C2)=NC2=C1N([Si](C)(C)C)C(N)=NC2=O | 4052.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #40 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C2)=NC2=C1N([Si](C)(C)C)C(N)=NC2=O | 6751.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #41 | C[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4235.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #41 | C[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 3957.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #41 | C[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 6797.6 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #5 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)C(=O)O[Si](C)(C)C | 4170.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #5 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)C(=O)O[Si](C)(C)C | 4006.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #5 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)C(=O)O[Si](C)(C)C | 6721.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O | 4433.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O | 4185.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O | 7273.7 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)=N2)N1[Si](C)(C)C | 4469.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)=N2)N1[Si](C)(C)C | 4165.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)=N2)N1[Si](C)(C)C | 7431.6 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #8 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C1)=N2 | 4346.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #8 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C1)=N2 | 4163.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #8 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C1)=N2 | 7128.6 | Standard polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #9 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4417.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #9 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4142.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TMS,isomer #9 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)[Si](C)(C)C)=N2)[NH]1 | 7409.4 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 4356.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 4117.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 6824.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #10 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 4044.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #10 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 3981.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #10 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 6264.2 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #11 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 4131.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #11 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 4036.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #11 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 6495.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #12 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4268.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #12 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4117.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #12 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 6807.9 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #13 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 4314.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #13 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 4175.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #13 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 6697.4 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #14 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O | 4406.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #14 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O | 4171.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #14 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O | 6758.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #15 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)C(=O)O | 4269.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #15 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)C(=O)O | 4197.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #15 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)C(=O)O | 6607.0 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #16 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)=N2)N1[Si](C)(C)C | 4304.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #16 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)=N2)N1[Si](C)(C)C | 4192.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #16 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)=N2)N1[Si](C)(C)C | 6593.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #17 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4337.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #17 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4112.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #17 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 6868.2 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #18 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4311.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #18 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4065.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #18 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 6993.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #19 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C1)[Si](C)(C)C)=N2 | 4227.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #19 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C1)[Si](C)(C)C)=N2 | 4125.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #19 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C1)[Si](C)(C)C)=N2 | 6625.4 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #2 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4376.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #2 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4059.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #2 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 7019.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #20 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C1)=N2 | 4204.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #20 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C1)=N2 | 4085.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #20 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C1)=N2 | 6714.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #21 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4233.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #21 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4036.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #21 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 6953.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #22 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4123.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #22 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3950.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #22 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 6480.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #23 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3996.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #23 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4005.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #23 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 6224.2 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #24 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 4085.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #24 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 4045.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #24 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 6466.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #25 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 4119.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #25 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 4098.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #25 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 6347.0 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #26 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4284.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #26 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4072.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #26 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 6766.2 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #27 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1 | 4307.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #27 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1 | 4117.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #27 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1 | 6629.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #28 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1 | 4389.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #28 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1 | 4116.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #28 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1 | 6691.4 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #29 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1 | 4258.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #29 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1 | 4156.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #29 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1 | 6542.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #3 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C1)=N2 | 4262.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #3 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C1)=N2 | 4064.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #3 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C1)=N2 | 6718.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #30 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4306.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #30 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4154.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #30 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 6529.9 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #31 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4332.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #31 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4064.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #31 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 6802.4 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #32 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4319.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #32 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4032.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #32 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 6953.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #33 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 4234.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #33 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 4090.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #33 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 6562.3 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #34 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C1)=N2 | 4209.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #34 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C1)=N2 | 4049.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #34 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C1)=N2 | 6676.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #35 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4246.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #35 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 3999.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #35 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 6912.4 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #36 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4133.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #36 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3921.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #36 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 6440.2 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #37 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3997.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #37 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3975.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #37 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 6186.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #38 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 4097.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #38 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 4012.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #38 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 6427.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #39 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1 | 4114.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #39 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1 | 4072.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #39 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1 | 6280.9 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #4 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4306.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #4 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4047.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #4 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 6993.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #40 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4360.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #40 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4272.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #40 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 6431.1 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #41 | C[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4413.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #41 | C[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4173.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #41 | C[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 6534.9 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #42 | C[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 4329.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #42 | C[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 4110.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #42 | C[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 6679.2 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #43 | C[Si](C)(C)N(CC1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4271.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #43 | C[Si](C)(C)N(CC1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4225.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #43 | C[Si](C)(C)N(CC1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 6425.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #44 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C2)=NC2=C1[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4192.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #44 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C2)=NC2=C1[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4181.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #44 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C2)=NC2=C1[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 6526.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #45 | C[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4255.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #45 | C[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4111.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #45 | C[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 6608.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #46 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4316.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #46 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4212.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #46 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 6386.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #47 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4255.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #47 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4159.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #47 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 6506.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #48 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4273.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #48 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4054.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #48 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 6764.3 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #49 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 4178.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #49 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 4105.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #49 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 6528.1 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #5 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)[NH]1 | 4317.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #5 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)[NH]1 | 4011.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #5 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)[NH]1 | 7103.2 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #50 | C[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4081.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #50 | C[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4082.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #50 | C[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 6259.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4152.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3878.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 6333.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #7 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 4224.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #7 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 3915.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #7 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 6675.2 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #8 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 4051.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #8 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 3946.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #8 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 6380.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #9 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 4187.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #9 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 3950.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TMS,isomer #9 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 6514.2 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4236.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4068.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 6430.6 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #10 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 4195.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #10 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 3975.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #10 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)[NH]1 | 6598.3 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #11 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4102.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #11 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3881.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #11 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 6197.2 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #12 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3957.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #12 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3924.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #12 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 5957.8 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #13 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 4036.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #13 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 3989.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #13 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C | 6208.3 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #14 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 4047.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #14 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 4022.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #14 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 6066.6 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #15 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4168.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #15 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4100.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #15 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 6275.6 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #16 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4246.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #16 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4099.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #16 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 6348.3 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #17 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 4142.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #17 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 4154.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #17 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 6215.3 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #18 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 4296.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #18 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 4135.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #18 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 6204.7 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #19 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)C(=O)O | 4196.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #19 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)C(=O)O | 4183.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #19 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)C(=O)O | 6117.1 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 4250.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 4085.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 6309.4 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #20 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O | 4271.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #20 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O | 4232.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #20 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O | 6095.7 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #21 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4208.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #21 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4149.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #21 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 6080.0 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #22 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4163.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #22 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4115.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #22 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 6196.8 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #23 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4196.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #23 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4008.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #23 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 6461.4 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #24 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 4102.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #24 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 4055.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #24 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 6233.0 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #25 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4015.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #25 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4044.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #25 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 6029.5 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #26 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4177.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #26 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4071.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #26 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 6245.4 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #27 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4256.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #27 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4074.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #27 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 6318.5 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #28 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 4142.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #28 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 4125.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #28 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 6186.8 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #29 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1 | 4283.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #29 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1 | 4100.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #29 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1 | 6152.1 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 4321.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 4075.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 6366.0 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #30 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1 | 4185.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #30 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1 | 4159.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #30 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1 | 6067.1 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #31 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1 | 4261.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #31 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1 | 4207.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #31 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1 | 6045.0 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #32 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4209.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #32 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4132.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #32 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 6030.0 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #33 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4171.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #33 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4093.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #33 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 6168.8 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #34 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4207.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #34 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 3990.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #34 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 6431.3 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #35 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 4098.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #35 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 4034.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #35 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 6204.4 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #36 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4026.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #36 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4030.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #36 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 5998.1 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #37 | C[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4286.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #37 | C[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4266.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #37 | C[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 5896.5 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #38 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C2)=NC2=C1N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4213.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #38 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C2)=NC2=C1N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 4226.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #38 | C[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C2)=NC2=C1N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O | 6028.5 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #39 | C[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4242.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #39 | C[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4103.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #39 | C[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 6133.0 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)C(=O)O[Si](C)(C)C | 4222.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)C(=O)O[Si](C)(C)C | 4115.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)C(=O)O[Si](C)(C)C | 6224.0 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #40 | C[Si](C)(C)N(CC1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4130.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #40 | C[Si](C)(C)N(CC1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4186.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #40 | C[Si](C)(C)N(CC1=NC2=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)N([Si](C)(C)C)C1)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 6038.7 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #41 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4177.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #41 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4145.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #41 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 6002.5 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #5 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4204.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #5 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4090.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #5 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 6201.6 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #6 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4247.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #6 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4000.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #6 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 6502.6 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4258.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 3994.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 6642.5 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #8 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 4166.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #8 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 4026.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #8 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 6250.7 | Standard polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #9 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C1)=N2 | 4148.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #9 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C1)=N2 | 4009.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,5TMS,isomer #9 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C1)=N2 | 6351.5 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4157.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4035.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #1 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 5927.6 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #10 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 4073.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #10 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 3983.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #10 | C[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C1)[Si](C)(C)C)=N2 | 5891.1 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #11 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3983.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #11 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 3977.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #11 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 5793.0 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #12 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4190.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #12 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4056.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #12 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 5828.3 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #13 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4091.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #13 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4152.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #13 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 5748.7 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #14 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4180.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #14 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4197.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #14 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 5730.9 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #15 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 4235.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #15 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 4214.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #15 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O | 5598.8 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #16 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4106.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #16 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4085.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #16 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 5712.4 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #17 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4191.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #17 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4045.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #17 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)[Si](C)(C)C | 5804.8 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #18 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4088.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #18 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 4136.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #18 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)[Si](C)(C)C | 5726.9 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #19 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4174.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #19 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4178.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #19 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 5709.4 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4227.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 4041.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #2 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)[Si](C)(C)C | 6001.7 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #20 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1 | 4216.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #20 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1 | 4204.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #20 | C[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1 | 5559.8 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #21 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4115.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #21 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4074.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #21 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 5691.0 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #22 | C[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4189.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #22 | C[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 4232.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #22 | C[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C)C1)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C)C=C1 | 5555.4 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 4122.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 4100.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #3 | C[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)C=C1)[Si](C)(C)C | 5875.5 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 4238.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 4036.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #4 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 5838.4 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #5 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 4159.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #5 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 4108.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #5 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)N([Si](C)(C)C)C2)[Si](C)(C)C)C=C1)C(=O)O[Si](C)(C)C | 5759.4 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 4216.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 4155.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #6 | C[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C)C2)N([Si](C)(C)C)C(N([Si](C)(C)C)[Si](C)(C)C)=NC3=O)C=C1)C(=O)O[Si](C)(C)C | 5727.7 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4152.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4052.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #7 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 5715.7 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #8 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4124.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #8 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 4045.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #8 | C[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)=N2)N1[Si](C)(C)C | 5849.5 | Standard polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #9 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 4171.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #9 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 3935.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,6TMS,isomer #9 | C[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C)C(=O)O[Si](C)(C)C)[Si](C)(C)C)C=C3)[Si](C)(C)C)=N2)N1[Si](C)(C)C | 6125.0 | Standard polar | 33892256 |
| Dihydrofolic acid,1TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)C(=O)O | 4888.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,1TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1 | 4861.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,1TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)[NH]1 | 4968.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,1TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1[NH]C(N)=NC2=O | 4770.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,1TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4863.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,1TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 4832.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,1TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)N1C(N)=NC(=O)C2=C1NCC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1)=N2 | 4794.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4920.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1 | 4860.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1 | 4785.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #12 | CC(C)(C)[Si](C)(C)N(C1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)[NH]1)[Si](C)(C)C(C)(C)C | 5001.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #13 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4977.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #14 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1)=N2 | 4894.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #15 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 5012.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #16 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 4951.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #17 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4795.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #18 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C2)=NC2=C1[NH]C(N)=NC2=O | 4750.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #19 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 4784.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)=N2)[NH]1 | 4983.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #20 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4875.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #21 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 4841.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #22 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 4826.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4874.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 4902.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)C(=O)O | 4882.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)C(=O)O | 4796.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 4971.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4873.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,2TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1 | 4884.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 4991.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 4613.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 7194.2 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 4932.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 4578.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 7150.4 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4843.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4399.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 6445.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #12 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4890.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #12 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4428.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #12 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 6663.2 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #13 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)[Si](C)(C)C(C)(C)C | 4769.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #13 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)[Si](C)(C)C(C)(C)C | 4475.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #13 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)[Si](C)(C)C(C)(C)C | 6488.6 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #14 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 4925.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #14 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 4470.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #14 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 6554.9 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #15 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 4818.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #15 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 4513.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #15 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 6412.6 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #16 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)C(=O)O | 4855.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #16 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)C(=O)O | 4588.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #16 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)C(=O)O | 6574.0 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #17 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1 | 5021.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #17 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1 | 4641.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #17 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1 | 6873.0 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #18 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 5024.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #18 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4575.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #18 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 6941.9 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #19 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C1)=N2 | 4904.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #19 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C1)=N2 | 4630.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #19 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C1)=N2 | 6817.1 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C(C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4905.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C(C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4396.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C(C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 6585.7 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #20 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 4978.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #20 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 4569.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #20 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 7037.9 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #21 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 4934.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #21 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 4547.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #21 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 7131.1 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #22 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4840.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #22 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4379.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #22 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 6422.5 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #23 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4890.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #23 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4404.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #23 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 6641.2 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #24 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)[Si](C)(C)C(C)(C)C | 4775.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #24 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)[Si](C)(C)C(C)(C)C | 4455.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #24 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)[Si](C)(C)C(C)(C)C | 6466.2 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #25 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1 | 4902.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #25 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1 | 4427.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #25 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1 | 6506.0 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #26 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1 | 4794.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #26 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1 | 4479.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #26 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1 | 6364.9 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #27 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1 | 4839.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #27 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1 | 4550.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #27 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1 | 6527.4 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #28 | CC(C)(C)[Si](C)(C)N(C1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 5101.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #28 | CC(C)(C)[Si](C)(C)N(C1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 4638.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #28 | CC(C)(C)[Si](C)(C)N(C1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 6842.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #29 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 4948.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #29 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 4697.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #29 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 6696.5 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4887.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4412.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 6471.4 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #30 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 5051.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #30 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4650.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #30 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 6760.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #31 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 4968.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #31 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 4616.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #31 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 6843.5 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #32 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4983.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #32 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4718.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #32 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 6638.1 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #33 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 5066.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #33 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 4580.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #33 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 6808.7 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #34 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4985.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #34 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4543.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #34 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 6906.2 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #35 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1)[Si](C)(C)C(C)(C)C)=N2 | 4934.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #35 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1)[Si](C)(C)C(C)(C)C)=N2 | 4645.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #35 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1)[Si](C)(C)C(C)(C)C)=N2 | 6731.2 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #36 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1)=N2 | 4874.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #36 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1)=N2 | 4618.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #36 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1)=N2 | 6796.6 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #37 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 4960.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #37 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 4543.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #37 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 6995.4 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #38 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4864.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #38 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4577.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #38 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 6417.3 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #39 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 4773.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #39 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 4485.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #39 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C([NH]C(N)=NC2=O)N([Si](C)(C)C(C)(C)C)C1)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 6348.0 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4934.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4450.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 6683.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #40 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C2)=NC2=C1N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 4814.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #40 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C2)=NC2=C1N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 4531.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #40 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C2)=NC2=C1N([Si](C)(C)C(C)(C)C)C(N)=NC2=O | 6516.3 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #41 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 4864.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #41 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 4406.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #41 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 6492.4 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4808.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4480.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 6510.2 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)C(=O)O | 5033.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)C(=O)O | 4706.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)C(=O)O | 6917.3 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 5035.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4628.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 6983.8 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C1)=N2 | 4915.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C1)=N2 | 4670.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C1)=N2 | 6859.5 | Standard polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 4990.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 4625.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,3TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 7082.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 5031.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4755.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 6528.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4820.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4535.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #10 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 6104.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4917.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4625.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #11 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 6282.9 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #12 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4974.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #12 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4746.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #12 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 6496.2 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #13 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 5071.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #13 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 4761.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #13 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 6410.4 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #14 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)C(=O)O | 5143.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #14 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)C(=O)O | 4743.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #14 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)C(=O)O | 6374.3 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #15 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)C(=O)O | 4982.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #15 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)C(=O)O | 4822.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #15 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)C(=O)O | 6351.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #16 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 5026.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #16 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4782.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #16 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 6295.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #17 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 5073.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #17 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 4647.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #17 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 6482.4 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #18 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 5022.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #18 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4631.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #18 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 6584.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #19 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C1)[Si](C)(C)C(C)(C)C)=N2 | 4953.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #19 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C1)[Si](C)(C)C(C)(C)C)=N2 | 4719.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #19 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)C=C1)[Si](C)(C)C(C)(C)C)=N2 | 6394.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 5069.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4639.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 6623.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #20 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1)=N2 | 4887.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #20 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1)=N2 | 4706.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #20 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1)=N2 | 6462.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #21 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 4949.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #21 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 4644.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #21 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 6665.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #22 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4930.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #22 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4489.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #22 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 6229.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #23 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4816.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #23 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4548.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #23 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 6086.4 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #24 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4903.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #24 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4621.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #24 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 6263.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #25 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 4963.4 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #25 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 4653.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #25 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O | 6166.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #26 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4975.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #26 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4723.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #26 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 6483.1 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #27 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1 | 5052.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #27 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1 | 4719.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #27 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1 | 6377.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #28 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1 | 5131.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #28 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1 | 4702.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #28 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1 | 6343.3 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #29 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1 | 4964.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #29 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1 | 4786.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #29 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(NCC2=NC3=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1 | 6320.2 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)C=C1)=N2 | 4936.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)C=C1)=N2 | 4700.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #3 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)C=C1)=N2 | 6482.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #30 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 5013.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #30 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4755.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #30 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CNC3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 6265.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #31 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 5059.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #31 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 4612.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #31 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 6450.0 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #32 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 5027.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #32 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4615.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #32 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 6572.1 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #33 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C)=N2 | 4938.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #33 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C)=N2 | 4692.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #33 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CN(C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C)=N2 | 6362.3 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #34 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C1)=N2 | 4883.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #34 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C1)=N2 | 4689.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #34 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CNC1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C1)=N2 | 6449.9 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #35 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 4944.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #35 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 4630.7 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #35 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 6652.0 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #36 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4928.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #36 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4480.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #36 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 6210.4 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #37 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4805.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #37 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4540.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #37 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(N(CC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 6067.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #38 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4903.1 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #38 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4609.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #38 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)N(C(=O)C1=CC=C(NCC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 6244.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #39 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1 | 4938.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #39 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1 | 4627.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #39 | CC(C)(C)[Si](C)(C)OC(=O)[C@H](CCC(=O)O)NC(=O)C1=CC=C(N(CC2=NC3=C(N([Si](C)(C)C(C)(C)C)C2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1 | 6130.9 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 4985.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 4654.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #4 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)[NH]1 | 6714.9 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #40 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 5092.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #40 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 4854.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #40 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C2)=NC2=C1N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 6159.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #41 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 5180.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #41 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4707.4 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #41 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 6201.1 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #42 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 5087.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #42 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 4687.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #42 | CC(C)(C)[Si](C)(C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC3=O)C=C1)[C@@H](CCC(=O)O)C(=O)O | 6318.0 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #43 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 5036.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #43 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 4807.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #43 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C([NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)N([Si](C)(C)C(C)(C)C)C1)C1=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C1 | 6216.3 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #44 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C2)=NC2=C1[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 4922.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #44 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C2)=NC2=C1[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 4796.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #44 | CC(C)(C)[Si](C)(C)N1CC(CNC2=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C2)=NC2=C1[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O | 6297.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #45 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 5023.6 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #45 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 4713.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #45 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(NC1)[NH]C(N([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 6346.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #46 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 5067.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #46 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 4772.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #46 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CN(C3=CC=C(C(=O)N[C@@H](CCC(=O)O)C(=O)O)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 6155.6 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #47 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4982.7 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #47 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 4745.3 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #47 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(N([Si](C)(C)C(C)(C)C)CC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)=N2)N1[Si](C)(C)C(C)(C)C | 6250.1 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #48 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 5036.3 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #48 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 4603.6 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #48 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CN(C3=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C3)[Si](C)(C)C(C)(C)C)=N2)N1[Si](C)(C)C(C)(C)C | 6414.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #49 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C)=N2 | 4916.8 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #49 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C)=N2 | 4706.5 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #49 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C([NH]1)N([Si](C)(C)C(C)(C)C)CC(CN(C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C)=N2 | 6339.1 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 4964.9 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 4659.8 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #5 | CC(C)(C)[Si](C)(C)NC1=NC(=O)C2=C(NCC(CNC3=CC=C(C(=O)N([C@@H](CCC(=O)O[Si](C)(C)C(C)(C)C)C(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)C=C3)=N2)[NH]1 | 6799.5 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #50 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 4920.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #50 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 4636.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #50 | CC(C)(C)[Si](C)(C)N(CC1=NC2=C(N([Si](C)(C)C(C)(C)C)C1)N([Si](C)(C)C(C)(C)C)C(N)=NC2=O)C1=CC=C(C(=O)N([C@@H](CCC(=O)O)C(=O)O)[Si](C)(C)C(C)(C)C)C=C1 | 6122.8 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C(C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4886.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C(C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 4456.1 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #6 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C(C)(C)C)N(C(=O)C1=CC=C(N(CC2=NC3=C(NC2)[NH]C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)[Si](C)(C)C(C)(C)C | 6140.2 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C(C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4959.2 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C(C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 4502.9 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #7 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C(C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)C=C1)[Si](C)(C)C(C)(C)C | 6377.7 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C(C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)[Si](C)(C)C(C)(C)C | 4804.0 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C(C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)[Si](C)(C)C(C)(C)C | 4534.0 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #8 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@@H](C(=O)O[Si](C)(C)C(C)(C)C)N(C(=O)C1=CC=C(NCC2=NC3=C([NH]C(N)=NC3=O)N([Si](C)(C)C(C)(C)C)C2)C=C1)[Si](C)(C)C(C)(C)C | 6203.9 | Standard polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4962.5 | Semi standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 4491.2 | Standard non polar | 33892256 |
| Dihydrofolic acid,4TBDMS,isomer #9 | CC(C)(C)[Si](C)(C)OC(=O)CC[C@H](NC(=O)C1=CC=C(N(CC2=NC3=C(NC2)N([Si](C)(C)C(C)(C)C)C(N)=NC3=O)[Si](C)(C)C(C)(C)C)C=C1)C(=O)O[Si](C)(C)C(C)(C)C | 6247.6 | Standard polar | 33892256 |